
Calothrix brevissima NIES-22
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Calotrichaceae; Calothrix; Calothrix brevissima
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
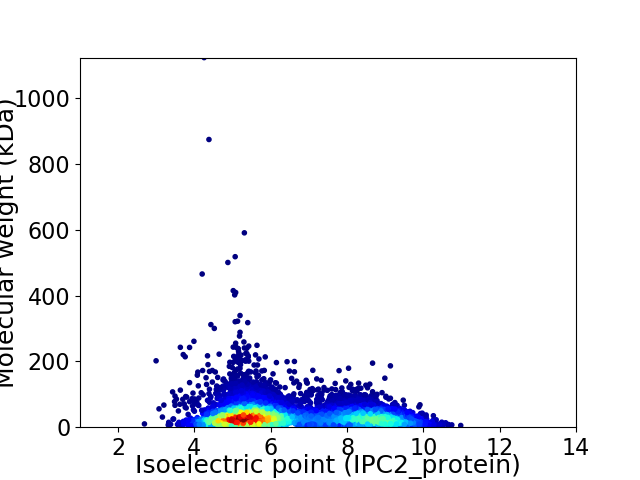
Virtual 2D-PAGE plot for 7229 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z4JWQ5|A0A1Z4JWQ5_9CYAN Cell wall hydrolase/autolysin OS=Calothrix brevissima NIES-22 OX=1973478 GN=NIES22_11700 PE=4 SV=1
MM1 pKa = 7.79SYY3 pKa = 8.12PTKK6 pKa = 10.56SFSIPNPVHH15 pKa = 6.6LVFIDD20 pKa = 3.99SAIQDD25 pKa = 3.3IQTLVEE31 pKa = 4.37GVVADD36 pKa = 3.84AQVVVLHH43 pKa = 6.38PEE45 pKa = 3.71VDD47 pKa = 3.34GVMQITQTLQNQPQIGTIHH66 pKa = 6.74LVAHH70 pKa = 7.17GYY72 pKa = 8.81PGCLCLGNAQLSLKK86 pKa = 8.98TLEE89 pKa = 4.44NYY91 pKa = 10.47APEE94 pKa = 3.92LRR96 pKa = 11.84IWRR99 pKa = 11.84EE100 pKa = 3.64VMSQQGQMLLYY111 pKa = 8.96GCHH114 pKa = 4.99VAAGEE119 pKa = 4.02MGAEE123 pKa = 4.81FVQKK127 pKa = 10.17LHH129 pKa = 6.12QLTGVKK135 pKa = 9.74IAASTTAIGNAAAGGNWEE153 pKa = 4.23LDD155 pKa = 3.46VRR157 pKa = 11.84LEE159 pKa = 3.96RR160 pKa = 11.84AATQVQPEE168 pKa = 4.41LPLAFRR174 pKa = 11.84AEE176 pKa = 3.8AMAAYY181 pKa = 10.11AGVLNTAPSSLIGSLPFDD199 pKa = 3.43VLKK202 pKa = 10.94NQWKK206 pKa = 8.1TLTINDD212 pKa = 4.58LLAGIIDD219 pKa = 4.63PDD221 pKa = 3.99NDD223 pKa = 3.88PLTITNLTASAGTILKK239 pKa = 10.18RR240 pKa = 11.84IDD242 pKa = 3.21GSYY245 pKa = 10.74SYY247 pKa = 10.52IPPVDD252 pKa = 4.08FFGGITFTYY261 pKa = 10.25DD262 pKa = 2.92VSDD265 pKa = 3.92GQNPPSQQTRR275 pKa = 11.84TVEE278 pKa = 3.95VLNQRR283 pKa = 11.84FPVLDD288 pKa = 3.58EE289 pKa = 4.86FGAGAAYY296 pKa = 10.54SNQAGVSDD304 pKa = 5.12LIKK307 pKa = 10.78VYY309 pKa = 10.17WDD311 pKa = 3.33YY312 pKa = 11.06DD313 pKa = 3.79TNGNVIATSRR323 pKa = 11.84NLAAAGNTTPSVLDD337 pKa = 3.45NDD339 pKa = 3.65IPSINFTWVSGNIPSQIGVLGFNTSNGGARR369 pKa = 11.84GIMWAVLGLVGKK381 pKa = 9.91SYY383 pKa = 10.54PDD385 pKa = 3.22EE386 pKa = 4.46IYY388 pKa = 10.51RR389 pKa = 11.84VGPYY393 pKa = 9.83RR394 pKa = 11.84DD395 pKa = 3.04TATNFFTSLQMDD407 pKa = 3.81VPGGVSGFINLSNPEE422 pKa = 4.16LKK424 pKa = 10.41FALEE428 pKa = 3.73QGKK431 pKa = 9.15PITEE435 pKa = 4.19SYY437 pKa = 10.64AYY439 pKa = 10.92LVMHH443 pKa = 6.98VDD445 pKa = 3.28NTVNHH450 pKa = 6.6PLISDD455 pKa = 3.29QDD457 pKa = 3.65VRR459 pKa = 11.84NGEE462 pKa = 4.08IVYY465 pKa = 8.4ATYY468 pKa = 10.37GYY470 pKa = 10.43YY471 pKa = 10.63NYY473 pKa = 8.96VTLSLEE479 pKa = 4.19RR480 pKa = 11.84NAPPVLTPKK489 pKa = 10.45SLSFQEE495 pKa = 4.93DD496 pKa = 3.71SGFQWIDD503 pKa = 2.99SRR505 pKa = 11.84LLEE508 pKa = 4.26GLQDD512 pKa = 3.63EE513 pKa = 4.75LAPGGFSSSPEE524 pKa = 3.87TFRR527 pKa = 11.84FGDD530 pKa = 3.5WFATYY535 pKa = 10.94GLTRR539 pKa = 11.84IDD541 pKa = 5.08GIPGVQALMQSIFYY555 pKa = 9.25TLNLYY560 pKa = 10.47DD561 pKa = 5.0LGGDD565 pKa = 3.31YY566 pKa = 9.91TAYY569 pKa = 10.65VLADD573 pKa = 3.7TGHH576 pKa = 7.17AAYY579 pKa = 10.05QHH581 pKa = 6.81LAQGATQTIVGSYY594 pKa = 7.92TVQDD598 pKa = 3.97DD599 pKa = 4.67FYY601 pKa = 9.69KK602 pKa = 10.72TSTAPLTVTFTGVNDD617 pKa = 3.82APVAVADD624 pKa = 4.26TARR627 pKa = 11.84GLPNTIIEE635 pKa = 4.53GSVATNDD642 pKa = 3.38SDD644 pKa = 4.41VDD646 pKa = 3.79DD647 pKa = 4.89GAILTYY653 pKa = 10.67SLSAPVDD660 pKa = 3.76GLTLNADD667 pKa = 3.55GSYY670 pKa = 10.92SFDD673 pKa = 3.32ATRR676 pKa = 11.84PAYY679 pKa = 10.46SSLPPGQILPVVVNYY694 pKa = 10.39RR695 pKa = 11.84VTDD698 pKa = 3.53EE699 pKa = 5.11FGAFSDD705 pKa = 3.85STLTLTLTLNSLPTGSPTVTLSDD728 pKa = 3.59TAEE731 pKa = 4.17DD732 pKa = 3.55TAITINTADD741 pKa = 4.21LLAGFSDD748 pKa = 4.27VEE750 pKa = 4.22GDD752 pKa = 3.64TLSVVGLTATNGSLVNNNDD771 pKa = 2.7GTYY774 pKa = 10.42TFTPTANYY782 pKa = 9.54NGNVTLTYY790 pKa = 10.46GVSDD794 pKa = 4.81DD795 pKa = 5.41KK796 pKa = 11.18EE797 pKa = 4.28TLANQTRR804 pKa = 11.84SFSVTPVNDD813 pKa = 3.72APVGSPTATLANTPEE828 pKa = 3.99DD829 pKa = 3.23TAIIINSTGLLIGFSDD845 pKa = 4.31VEE847 pKa = 4.27GDD849 pKa = 3.64TLSVVGLTATNGSLVNNNNGTYY871 pKa = 10.16TFTPTANYY879 pKa = 9.54NGNVTLTYY887 pKa = 10.04GVSDD891 pKa = 3.58GTVTLTGQTRR901 pKa = 11.84SFSVTPVNDD910 pKa = 3.72APVGSPTATLANTRR924 pKa = 11.84EE925 pKa = 4.01DD926 pKa = 3.18TAIIINSADD935 pKa = 3.56LLAGFSDD942 pKa = 3.52VDD944 pKa = 3.8GNNTLSVVGLTATNGSLVNNNNGTYY969 pKa = 10.16TFTPTANYY977 pKa = 9.54NGNVTLTYY985 pKa = 10.04GVSDD989 pKa = 3.69GTVTLAGQTRR999 pKa = 11.84SFSVTPVNDD1008 pKa = 3.44APVAQSPPATTYY1020 pKa = 10.47QAINTPYY1027 pKa = 10.24QAIDD1031 pKa = 3.82LVPGGSGVVSILSNDD1046 pKa = 3.84DD1047 pKa = 4.17DD1048 pKa = 4.89NFAQINLGGASFNFYY1063 pKa = 11.21GNTYY1067 pKa = 10.03NSLFASSNGLITFGTGEE1084 pKa = 4.09TGFFNTDD1091 pKa = 2.98LSSGLGGLTNEE1102 pKa = 4.19IRR1104 pKa = 11.84QAAIAVFWDD1113 pKa = 3.71DD1114 pKa = 5.68LSTQGTTANQVLYY1127 pKa = 10.55RR1128 pKa = 11.84IQDD1131 pKa = 3.34NQLIVEE1137 pKa = 4.41WKK1139 pKa = 8.83EE1140 pKa = 3.59VFIRR1144 pKa = 11.84GSGSGNPITFQAILQLNTGLARR1166 pKa = 11.84GNITFNYY1173 pKa = 9.51VDD1175 pKa = 4.76LDD1177 pKa = 4.02AGNVSYY1183 pKa = 11.67NNGASATVGIKK1194 pKa = 10.28NATGSIGSPVLVSLNNGSNPNVRR1217 pKa = 11.84SGQAITIFEE1226 pKa = 4.44VGPLGYY1232 pKa = 7.51TTEE1235 pKa = 4.12DD1236 pKa = 2.92TAIIINSADD1245 pKa = 3.76LLTSFSDD1252 pKa = 3.93VEE1254 pKa = 4.3GDD1256 pKa = 3.51TLSVVNLTATNGSLVNNNNGTYY1278 pKa = 10.16TFTPTANFNGKK1289 pKa = 9.63VNLTYY1294 pKa = 10.55GVSDD1298 pKa = 5.56GIDD1301 pKa = 3.27TLSGQVQTFPVIPVNDD1317 pKa = 4.06APVGSPTATLANTRR1331 pKa = 11.84EE1332 pKa = 4.01DD1333 pKa = 3.18TAIIINSADD1342 pKa = 3.56LLAGFSDD1349 pKa = 4.27VEE1351 pKa = 4.22GDD1353 pKa = 3.64TLSVVGLTATNGSLVNNNNGTYY1375 pKa = 10.16TFTPTANFKK1384 pKa = 9.72GTVTLTYY1391 pKa = 9.86GVSDD1395 pKa = 3.69GTVTLAGQTRR1405 pKa = 11.84SFSVTPINDD1414 pKa = 3.58APVGSPTATLSDD1426 pKa = 3.95TPEE1429 pKa = 3.95DD1430 pKa = 3.22TAIIINSADD1439 pKa = 3.56LLAGFSDD1446 pKa = 4.18ANGDD1450 pKa = 3.81TLSVVGLTADD1460 pKa = 3.47NGSLVINNDD1469 pKa = 2.47GTYY1472 pKa = 10.2TFNPTANFNGAVNLTYY1488 pKa = 10.81GVTDD1492 pKa = 4.03GTATLASQTRR1502 pKa = 11.84SFNVTAINDD1511 pKa = 3.9APVGSPTATLANTSEE1526 pKa = 4.05DD1527 pKa = 3.33TAINITAADD1536 pKa = 4.13LLEE1539 pKa = 4.67GFSDD1543 pKa = 3.67VDD1545 pKa = 3.46GDD1547 pKa = 3.89NLSVVGLTADD1557 pKa = 3.47NGSLVINNDD1566 pKa = 2.47GTYY1569 pKa = 10.2TFNPTANFNGAVNLTYY1585 pKa = 10.6SVSDD1589 pKa = 3.45GTASLTGRR1597 pKa = 11.84TRR1599 pKa = 11.84TFTVTAVNDD1608 pKa = 3.96APVGSPTATLSNTAEE1623 pKa = 4.15DD1624 pKa = 3.6TAITINSSNLLEE1636 pKa = 4.71GFSDD1640 pKa = 3.57VDD1642 pKa = 3.46GDD1644 pKa = 3.89NLSVVGLTADD1654 pKa = 3.47NGSLVINNDD1663 pKa = 2.47GTYY1666 pKa = 10.2TFNPTANFNGAVNLTYY1682 pKa = 10.6SVSDD1686 pKa = 3.45GTASLTGRR1694 pKa = 11.84TRR1696 pKa = 11.84TFTVTAVNDD1705 pKa = 3.96APVGSPTATLSNTAEE1720 pKa = 4.15DD1721 pKa = 3.6TAITINSSNLLAGFSDD1737 pKa = 3.52VDD1739 pKa = 3.59GDD1741 pKa = 3.89NLSVVGLTATNGSLVNNNNGTYY1763 pKa = 10.16TFTPTANFNGTVTLTYY1779 pKa = 10.83DD1780 pKa = 3.56VTDD1783 pKa = 4.03GTATLAGQTRR1793 pKa = 11.84SFNITAVNDD1802 pKa = 3.82APTGSPTATLSNTAEE1817 pKa = 3.93DD1818 pKa = 3.6TAINITVANLLAGFSDD1834 pKa = 3.6VDD1836 pKa = 3.63GDD1838 pKa = 3.99TLSVVNLTANNGSLVNNNNGTYY1860 pKa = 10.16TFTPTANFNGTVNLTYY1876 pKa = 10.48GVSDD1880 pKa = 3.8GTATFAGQTRR1890 pKa = 11.84SFAVTPVNDD1899 pKa = 4.32APTGSPTAALSNTPEE1914 pKa = 3.92DD1915 pKa = 3.36TAINITASSLLAGFTDD1931 pKa = 3.35VDD1933 pKa = 3.95GDD1935 pKa = 3.96TLSVVNLTATNGALVNNNNGTYY1957 pKa = 10.16TFTPTANFNGTVNLTYY1973 pKa = 10.48GVSDD1977 pKa = 3.82GTATLAGQTRR1987 pKa = 11.84SFAVTPVNDD1996 pKa = 4.32APTGSPTAALSNTPEE2011 pKa = 3.77EE2012 pKa = 3.97TAINITASSLLAGFTDD2028 pKa = 3.35VDD2030 pKa = 3.95GDD2032 pKa = 3.96TLSVVNLTATNGALVNNNNGTYY2054 pKa = 10.19TFTPNTNFNGTVNLTYY2070 pKa = 10.3GVSDD2074 pKa = 3.59GTVTLAGQTRR2084 pKa = 11.84SFTVTPVNDD2093 pKa = 3.83APVGNPDD2100 pKa = 3.48TVSTTRR2106 pKa = 11.84NTAVNIAVSTLLANDD2121 pKa = 3.76TDD2123 pKa = 3.63IDD2125 pKa = 4.03TPLASLRR2132 pKa = 11.84ITAVSGATNGTVSLNNKK2149 pKa = 7.01GTPTNFSDD2157 pKa = 5.26DD2158 pKa = 4.54FITFTPQNNFTGNATFNYY2176 pKa = 8.06TISDD2180 pKa = 3.69GSLTSTATVAVSVGLSLNGSNGSNILTGSDD2210 pKa = 3.68FSDD2213 pKa = 5.16LINGNDD2219 pKa = 4.27GDD2221 pKa = 5.49DD2222 pKa = 3.27ILYY2225 pKa = 10.78GNNGNDD2231 pKa = 3.47TLIGGNGLDD2240 pKa = 4.08LLLGGEE2246 pKa = 4.96GNDD2249 pKa = 3.78SLNGGGGADD2258 pKa = 3.36ALYY2261 pKa = 10.54GGKK2264 pKa = 10.8GNDD2267 pKa = 3.47TLTGGSGIDD2276 pKa = 3.05IFAFAQGDD2284 pKa = 4.02GSDD2287 pKa = 4.12TITDD2291 pKa = 3.91FTDD2294 pKa = 5.05GIDD2297 pKa = 3.57WIGLVGLRR2305 pKa = 11.84FNQLSISGNGGNTTIRR2321 pKa = 11.84YY2322 pKa = 9.55GNEE2325 pKa = 3.3TLATLTGVSANLITAADD2342 pKa = 5.11FISVV2346 pKa = 3.3
MM1 pKa = 7.79SYY3 pKa = 8.12PTKK6 pKa = 10.56SFSIPNPVHH15 pKa = 6.6LVFIDD20 pKa = 3.99SAIQDD25 pKa = 3.3IQTLVEE31 pKa = 4.37GVVADD36 pKa = 3.84AQVVVLHH43 pKa = 6.38PEE45 pKa = 3.71VDD47 pKa = 3.34GVMQITQTLQNQPQIGTIHH66 pKa = 6.74LVAHH70 pKa = 7.17GYY72 pKa = 8.81PGCLCLGNAQLSLKK86 pKa = 8.98TLEE89 pKa = 4.44NYY91 pKa = 10.47APEE94 pKa = 3.92LRR96 pKa = 11.84IWRR99 pKa = 11.84EE100 pKa = 3.64VMSQQGQMLLYY111 pKa = 8.96GCHH114 pKa = 4.99VAAGEE119 pKa = 4.02MGAEE123 pKa = 4.81FVQKK127 pKa = 10.17LHH129 pKa = 6.12QLTGVKK135 pKa = 9.74IAASTTAIGNAAAGGNWEE153 pKa = 4.23LDD155 pKa = 3.46VRR157 pKa = 11.84LEE159 pKa = 3.96RR160 pKa = 11.84AATQVQPEE168 pKa = 4.41LPLAFRR174 pKa = 11.84AEE176 pKa = 3.8AMAAYY181 pKa = 10.11AGVLNTAPSSLIGSLPFDD199 pKa = 3.43VLKK202 pKa = 10.94NQWKK206 pKa = 8.1TLTINDD212 pKa = 4.58LLAGIIDD219 pKa = 4.63PDD221 pKa = 3.99NDD223 pKa = 3.88PLTITNLTASAGTILKK239 pKa = 10.18RR240 pKa = 11.84IDD242 pKa = 3.21GSYY245 pKa = 10.74SYY247 pKa = 10.52IPPVDD252 pKa = 4.08FFGGITFTYY261 pKa = 10.25DD262 pKa = 2.92VSDD265 pKa = 3.92GQNPPSQQTRR275 pKa = 11.84TVEE278 pKa = 3.95VLNQRR283 pKa = 11.84FPVLDD288 pKa = 3.58EE289 pKa = 4.86FGAGAAYY296 pKa = 10.54SNQAGVSDD304 pKa = 5.12LIKK307 pKa = 10.78VYY309 pKa = 10.17WDD311 pKa = 3.33YY312 pKa = 11.06DD313 pKa = 3.79TNGNVIATSRR323 pKa = 11.84NLAAAGNTTPSVLDD337 pKa = 3.45NDD339 pKa = 3.65IPSINFTWVSGNIPSQIGVLGFNTSNGGARR369 pKa = 11.84GIMWAVLGLVGKK381 pKa = 9.91SYY383 pKa = 10.54PDD385 pKa = 3.22EE386 pKa = 4.46IYY388 pKa = 10.51RR389 pKa = 11.84VGPYY393 pKa = 9.83RR394 pKa = 11.84DD395 pKa = 3.04TATNFFTSLQMDD407 pKa = 3.81VPGGVSGFINLSNPEE422 pKa = 4.16LKK424 pKa = 10.41FALEE428 pKa = 3.73QGKK431 pKa = 9.15PITEE435 pKa = 4.19SYY437 pKa = 10.64AYY439 pKa = 10.92LVMHH443 pKa = 6.98VDD445 pKa = 3.28NTVNHH450 pKa = 6.6PLISDD455 pKa = 3.29QDD457 pKa = 3.65VRR459 pKa = 11.84NGEE462 pKa = 4.08IVYY465 pKa = 8.4ATYY468 pKa = 10.37GYY470 pKa = 10.43YY471 pKa = 10.63NYY473 pKa = 8.96VTLSLEE479 pKa = 4.19RR480 pKa = 11.84NAPPVLTPKK489 pKa = 10.45SLSFQEE495 pKa = 4.93DD496 pKa = 3.71SGFQWIDD503 pKa = 2.99SRR505 pKa = 11.84LLEE508 pKa = 4.26GLQDD512 pKa = 3.63EE513 pKa = 4.75LAPGGFSSSPEE524 pKa = 3.87TFRR527 pKa = 11.84FGDD530 pKa = 3.5WFATYY535 pKa = 10.94GLTRR539 pKa = 11.84IDD541 pKa = 5.08GIPGVQALMQSIFYY555 pKa = 9.25TLNLYY560 pKa = 10.47DD561 pKa = 5.0LGGDD565 pKa = 3.31YY566 pKa = 9.91TAYY569 pKa = 10.65VLADD573 pKa = 3.7TGHH576 pKa = 7.17AAYY579 pKa = 10.05QHH581 pKa = 6.81LAQGATQTIVGSYY594 pKa = 7.92TVQDD598 pKa = 3.97DD599 pKa = 4.67FYY601 pKa = 9.69KK602 pKa = 10.72TSTAPLTVTFTGVNDD617 pKa = 3.82APVAVADD624 pKa = 4.26TARR627 pKa = 11.84GLPNTIIEE635 pKa = 4.53GSVATNDD642 pKa = 3.38SDD644 pKa = 4.41VDD646 pKa = 3.79DD647 pKa = 4.89GAILTYY653 pKa = 10.67SLSAPVDD660 pKa = 3.76GLTLNADD667 pKa = 3.55GSYY670 pKa = 10.92SFDD673 pKa = 3.32ATRR676 pKa = 11.84PAYY679 pKa = 10.46SSLPPGQILPVVVNYY694 pKa = 10.39RR695 pKa = 11.84VTDD698 pKa = 3.53EE699 pKa = 5.11FGAFSDD705 pKa = 3.85STLTLTLTLNSLPTGSPTVTLSDD728 pKa = 3.59TAEE731 pKa = 4.17DD732 pKa = 3.55TAITINTADD741 pKa = 4.21LLAGFSDD748 pKa = 4.27VEE750 pKa = 4.22GDD752 pKa = 3.64TLSVVGLTATNGSLVNNNDD771 pKa = 2.7GTYY774 pKa = 10.42TFTPTANYY782 pKa = 9.54NGNVTLTYY790 pKa = 10.46GVSDD794 pKa = 4.81DD795 pKa = 5.41KK796 pKa = 11.18EE797 pKa = 4.28TLANQTRR804 pKa = 11.84SFSVTPVNDD813 pKa = 3.72APVGSPTATLANTPEE828 pKa = 3.99DD829 pKa = 3.23TAIIINSTGLLIGFSDD845 pKa = 4.31VEE847 pKa = 4.27GDD849 pKa = 3.64TLSVVGLTATNGSLVNNNNGTYY871 pKa = 10.16TFTPTANYY879 pKa = 9.54NGNVTLTYY887 pKa = 10.04GVSDD891 pKa = 3.58GTVTLTGQTRR901 pKa = 11.84SFSVTPVNDD910 pKa = 3.72APVGSPTATLANTRR924 pKa = 11.84EE925 pKa = 4.01DD926 pKa = 3.18TAIIINSADD935 pKa = 3.56LLAGFSDD942 pKa = 3.52VDD944 pKa = 3.8GNNTLSVVGLTATNGSLVNNNNGTYY969 pKa = 10.16TFTPTANYY977 pKa = 9.54NGNVTLTYY985 pKa = 10.04GVSDD989 pKa = 3.69GTVTLAGQTRR999 pKa = 11.84SFSVTPVNDD1008 pKa = 3.44APVAQSPPATTYY1020 pKa = 10.47QAINTPYY1027 pKa = 10.24QAIDD1031 pKa = 3.82LVPGGSGVVSILSNDD1046 pKa = 3.84DD1047 pKa = 4.17DD1048 pKa = 4.89NFAQINLGGASFNFYY1063 pKa = 11.21GNTYY1067 pKa = 10.03NSLFASSNGLITFGTGEE1084 pKa = 4.09TGFFNTDD1091 pKa = 2.98LSSGLGGLTNEE1102 pKa = 4.19IRR1104 pKa = 11.84QAAIAVFWDD1113 pKa = 3.71DD1114 pKa = 5.68LSTQGTTANQVLYY1127 pKa = 10.55RR1128 pKa = 11.84IQDD1131 pKa = 3.34NQLIVEE1137 pKa = 4.41WKK1139 pKa = 8.83EE1140 pKa = 3.59VFIRR1144 pKa = 11.84GSGSGNPITFQAILQLNTGLARR1166 pKa = 11.84GNITFNYY1173 pKa = 9.51VDD1175 pKa = 4.76LDD1177 pKa = 4.02AGNVSYY1183 pKa = 11.67NNGASATVGIKK1194 pKa = 10.28NATGSIGSPVLVSLNNGSNPNVRR1217 pKa = 11.84SGQAITIFEE1226 pKa = 4.44VGPLGYY1232 pKa = 7.51TTEE1235 pKa = 4.12DD1236 pKa = 2.92TAIIINSADD1245 pKa = 3.76LLTSFSDD1252 pKa = 3.93VEE1254 pKa = 4.3GDD1256 pKa = 3.51TLSVVNLTATNGSLVNNNNGTYY1278 pKa = 10.16TFTPTANFNGKK1289 pKa = 9.63VNLTYY1294 pKa = 10.55GVSDD1298 pKa = 5.56GIDD1301 pKa = 3.27TLSGQVQTFPVIPVNDD1317 pKa = 4.06APVGSPTATLANTRR1331 pKa = 11.84EE1332 pKa = 4.01DD1333 pKa = 3.18TAIIINSADD1342 pKa = 3.56LLAGFSDD1349 pKa = 4.27VEE1351 pKa = 4.22GDD1353 pKa = 3.64TLSVVGLTATNGSLVNNNNGTYY1375 pKa = 10.16TFTPTANFKK1384 pKa = 9.72GTVTLTYY1391 pKa = 9.86GVSDD1395 pKa = 3.69GTVTLAGQTRR1405 pKa = 11.84SFSVTPINDD1414 pKa = 3.58APVGSPTATLSDD1426 pKa = 3.95TPEE1429 pKa = 3.95DD1430 pKa = 3.22TAIIINSADD1439 pKa = 3.56LLAGFSDD1446 pKa = 4.18ANGDD1450 pKa = 3.81TLSVVGLTADD1460 pKa = 3.47NGSLVINNDD1469 pKa = 2.47GTYY1472 pKa = 10.2TFNPTANFNGAVNLTYY1488 pKa = 10.81GVTDD1492 pKa = 4.03GTATLASQTRR1502 pKa = 11.84SFNVTAINDD1511 pKa = 3.9APVGSPTATLANTSEE1526 pKa = 4.05DD1527 pKa = 3.33TAINITAADD1536 pKa = 4.13LLEE1539 pKa = 4.67GFSDD1543 pKa = 3.67VDD1545 pKa = 3.46GDD1547 pKa = 3.89NLSVVGLTADD1557 pKa = 3.47NGSLVINNDD1566 pKa = 2.47GTYY1569 pKa = 10.2TFNPTANFNGAVNLTYY1585 pKa = 10.6SVSDD1589 pKa = 3.45GTASLTGRR1597 pKa = 11.84TRR1599 pKa = 11.84TFTVTAVNDD1608 pKa = 3.96APVGSPTATLSNTAEE1623 pKa = 4.15DD1624 pKa = 3.6TAITINSSNLLEE1636 pKa = 4.71GFSDD1640 pKa = 3.57VDD1642 pKa = 3.46GDD1644 pKa = 3.89NLSVVGLTADD1654 pKa = 3.47NGSLVINNDD1663 pKa = 2.47GTYY1666 pKa = 10.2TFNPTANFNGAVNLTYY1682 pKa = 10.6SVSDD1686 pKa = 3.45GTASLTGRR1694 pKa = 11.84TRR1696 pKa = 11.84TFTVTAVNDD1705 pKa = 3.96APVGSPTATLSNTAEE1720 pKa = 4.15DD1721 pKa = 3.6TAITINSSNLLAGFSDD1737 pKa = 3.52VDD1739 pKa = 3.59GDD1741 pKa = 3.89NLSVVGLTATNGSLVNNNNGTYY1763 pKa = 10.16TFTPTANFNGTVTLTYY1779 pKa = 10.83DD1780 pKa = 3.56VTDD1783 pKa = 4.03GTATLAGQTRR1793 pKa = 11.84SFNITAVNDD1802 pKa = 3.82APTGSPTATLSNTAEE1817 pKa = 3.93DD1818 pKa = 3.6TAINITVANLLAGFSDD1834 pKa = 3.6VDD1836 pKa = 3.63GDD1838 pKa = 3.99TLSVVNLTANNGSLVNNNNGTYY1860 pKa = 10.16TFTPTANFNGTVNLTYY1876 pKa = 10.48GVSDD1880 pKa = 3.8GTATFAGQTRR1890 pKa = 11.84SFAVTPVNDD1899 pKa = 4.32APTGSPTAALSNTPEE1914 pKa = 3.92DD1915 pKa = 3.36TAINITASSLLAGFTDD1931 pKa = 3.35VDD1933 pKa = 3.95GDD1935 pKa = 3.96TLSVVNLTATNGALVNNNNGTYY1957 pKa = 10.16TFTPTANFNGTVNLTYY1973 pKa = 10.48GVSDD1977 pKa = 3.82GTATLAGQTRR1987 pKa = 11.84SFAVTPVNDD1996 pKa = 4.32APTGSPTAALSNTPEE2011 pKa = 3.77EE2012 pKa = 3.97TAINITASSLLAGFTDD2028 pKa = 3.35VDD2030 pKa = 3.95GDD2032 pKa = 3.96TLSVVNLTATNGALVNNNNGTYY2054 pKa = 10.19TFTPNTNFNGTVNLTYY2070 pKa = 10.3GVSDD2074 pKa = 3.59GTVTLAGQTRR2084 pKa = 11.84SFTVTPVNDD2093 pKa = 3.83APVGNPDD2100 pKa = 3.48TVSTTRR2106 pKa = 11.84NTAVNIAVSTLLANDD2121 pKa = 3.76TDD2123 pKa = 3.63IDD2125 pKa = 4.03TPLASLRR2132 pKa = 11.84ITAVSGATNGTVSLNNKK2149 pKa = 7.01GTPTNFSDD2157 pKa = 5.26DD2158 pKa = 4.54FITFTPQNNFTGNATFNYY2176 pKa = 8.06TISDD2180 pKa = 3.69GSLTSTATVAVSVGLSLNGSNGSNILTGSDD2210 pKa = 3.68FSDD2213 pKa = 5.16LINGNDD2219 pKa = 4.27GDD2221 pKa = 5.49DD2222 pKa = 3.27ILYY2225 pKa = 10.78GNNGNDD2231 pKa = 3.47TLIGGNGLDD2240 pKa = 4.08LLLGGEE2246 pKa = 4.96GNDD2249 pKa = 3.78SLNGGGGADD2258 pKa = 3.36ALYY2261 pKa = 10.54GGKK2264 pKa = 10.8GNDD2267 pKa = 3.47TLTGGSGIDD2276 pKa = 3.05IFAFAQGDD2284 pKa = 4.02GSDD2287 pKa = 4.12TITDD2291 pKa = 3.91FTDD2294 pKa = 5.05GIDD2297 pKa = 3.57WIGLVGLRR2305 pKa = 11.84FNQLSISGNGGNTTIRR2321 pKa = 11.84YY2322 pKa = 9.55GNEE2325 pKa = 3.3TLATLTGVSANLITAADD2342 pKa = 5.11FISVV2346 pKa = 3.3
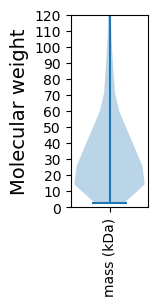
Molecular weight: 242.76 kDa
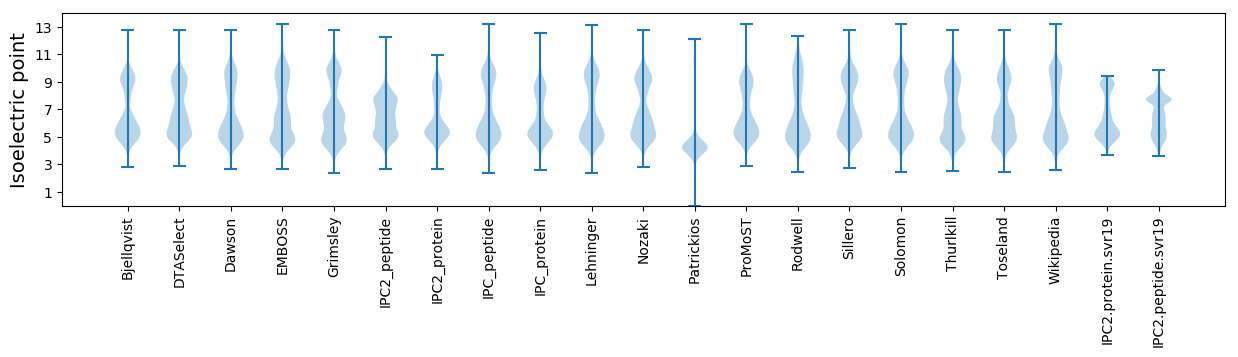
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z4JYW6|A0A1Z4JYW6_9CYAN CMP/dCMP deaminase zinc-binding protein OS=Calothrix brevissima NIES-22 OX=1973478 GN=NIES22_19000 PE=4 SV=1
MM1 pKa = 7.08QRR3 pKa = 11.84TLGGTNRR10 pKa = 11.84KK11 pKa = 9.1RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TPDD26 pKa = 3.17GRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.01KK37 pKa = 9.6GRR39 pKa = 11.84HH40 pKa = 5.0RR41 pKa = 11.84LSVV44 pKa = 3.12
MM1 pKa = 7.08QRR3 pKa = 11.84TLGGTNRR10 pKa = 11.84KK11 pKa = 9.1RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TPDD26 pKa = 3.17GRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.01KK37 pKa = 9.6GRR39 pKa = 11.84HH40 pKa = 5.0RR41 pKa = 11.84LSVV44 pKa = 3.12
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2460895 |
29 |
10429 |
340.4 |
37.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.191 ± 0.033 | 0.931 ± 0.011 |
4.88 ± 0.028 | 6.038 ± 0.03 |
3.896 ± 0.018 | 6.518 ± 0.04 |
1.771 ± 0.014 | 7.076 ± 0.026 |
4.993 ± 0.03 | 10.886 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.696 ± 0.017 | 4.868 ± 0.039 |
4.6 ± 0.024 | 5.658 ± 0.031 |
4.818 ± 0.027 | 6.467 ± 0.024 |
5.79 ± 0.034 | 6.398 ± 0.023 |
1.394 ± 0.014 | 3.13 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
