
Lishi spider virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Chuviridae; Scarabeuvirus; Scarabeuvirus lishiense
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
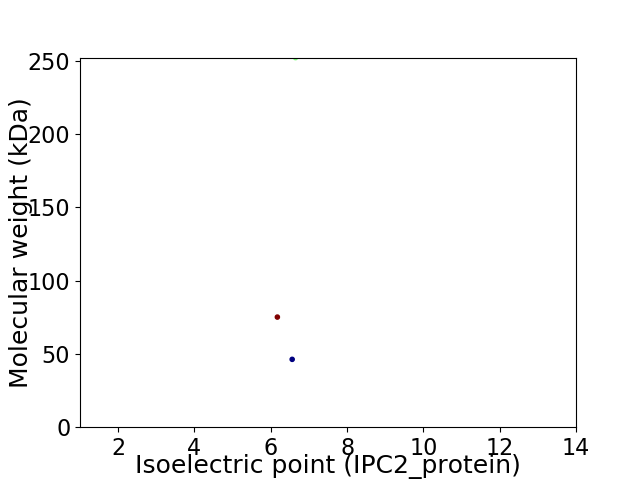
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
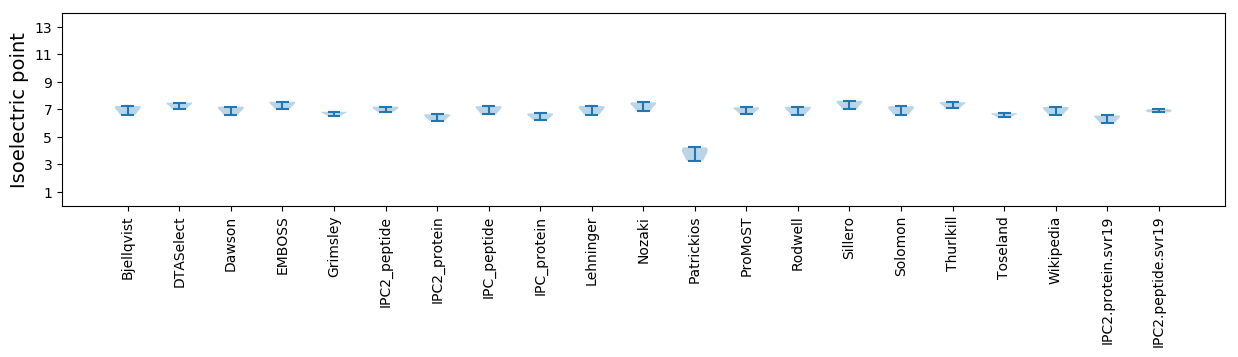
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KJV2|A0A0B5KJV2_9VIRU Putative nucleoprotein OS=Lishi spider virus 1 OX=1608057 GN=N PE=4 SV=1
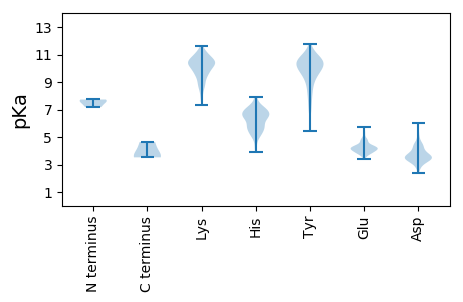
MM1 pKa = 7.73LILEE5 pKa = 4.58SNMVNYY11 pKa = 9.2MIKK14 pKa = 10.59LLFLPTCYY22 pKa = 10.59GFLAFDD28 pKa = 4.41CSNPNINYY36 pKa = 8.29TLVNTRR42 pKa = 11.84EE43 pKa = 3.98VEE45 pKa = 3.98EE46 pKa = 5.25CIISTPEE53 pKa = 3.44IVEE56 pKa = 4.21EE57 pKa = 4.11KK58 pKa = 11.07VSIQLLQLNSIAYY71 pKa = 9.1AKK73 pKa = 10.42YY74 pKa = 9.03VSCKK78 pKa = 9.65VSMNRR83 pKa = 11.84IISYY87 pKa = 10.55CGMNSHH93 pKa = 6.34VSMVHH98 pKa = 5.58NGYY101 pKa = 11.1ASFIVPITNTEE112 pKa = 3.75CWNIHH117 pKa = 5.22RR118 pKa = 11.84DD119 pKa = 3.34RR120 pKa = 11.84RR121 pKa = 11.84YY122 pKa = 9.72ILPNLSVVSNLKK134 pKa = 10.02PNSTIEE140 pKa = 4.04VSSVLSGKK148 pKa = 10.02LFSDD152 pKa = 4.94GKK154 pKa = 11.04CEE156 pKa = 4.04GGSYY160 pKa = 9.4NDD162 pKa = 5.25KK163 pKa = 9.92YY164 pKa = 11.3GSWEE168 pKa = 4.11SVIVQAIYY176 pKa = 10.35KK177 pKa = 9.27IQLTEE182 pKa = 4.61GYY184 pKa = 7.35TQYY187 pKa = 11.55DD188 pKa = 3.38RR189 pKa = 11.84DD190 pKa = 3.68TNYY193 pKa = 9.75IHH195 pKa = 7.21LPYY198 pKa = 10.27GINCIYY204 pKa = 10.95DD205 pKa = 4.3RR206 pKa = 11.84IDD208 pKa = 3.79CVHH211 pKa = 7.42PDD213 pKa = 2.9IGMTTWDD220 pKa = 3.46VFPTHH225 pKa = 6.55EE226 pKa = 4.79CDD228 pKa = 4.28DD229 pKa = 3.8NRR231 pKa = 11.84HH232 pKa = 5.64LVLFDD237 pKa = 3.65GFASKK242 pKa = 7.73TTTQSLTGEE251 pKa = 3.99NMKK254 pKa = 10.33VVTYY258 pKa = 9.0MVTSSTRR265 pKa = 11.84AFALSVTTRR274 pKa = 11.84YY275 pKa = 9.63DD276 pKa = 3.22KK277 pKa = 11.34CSMEE281 pKa = 4.3VWRR284 pKa = 11.84TEE286 pKa = 3.86HH287 pKa = 6.71PRR289 pKa = 11.84LLILVVNQQWDD300 pKa = 4.03KK301 pKa = 11.13IKK303 pKa = 11.11SNSIDD308 pKa = 3.34ISSLDD313 pKa = 3.29LMAYY317 pKa = 10.19VNAKK321 pKa = 10.37LIMIQASVASNVEE334 pKa = 4.03SLYY337 pKa = 9.36QTLDD341 pKa = 3.01YY342 pKa = 9.79RR343 pKa = 11.84RR344 pKa = 11.84CMAEE348 pKa = 3.86RR349 pKa = 11.84KK350 pKa = 9.46SLRR353 pKa = 11.84NQLALASLSPQEE365 pKa = 3.87FAYY368 pKa = 10.65LYY370 pKa = 8.9MGKK373 pKa = 9.26PGYY376 pKa = 9.04TSLPMGEE383 pKa = 3.9VSYY386 pKa = 10.52IIQCNPVEE394 pKa = 4.34VKK396 pKa = 10.24LRR398 pKa = 11.84KK399 pKa = 8.07TDD401 pKa = 3.05KK402 pKa = 10.43CYY404 pKa = 10.82HH405 pKa = 6.38EE406 pKa = 5.62LPISLNNQSFYY417 pKa = 10.58MSPKK421 pKa = 8.62TRR423 pKa = 11.84LIQSQGTEE431 pKa = 4.18VEE433 pKa = 4.86CNHH436 pKa = 6.61LMLPKK441 pKa = 10.33FLLGKK446 pKa = 9.57NWYY449 pKa = 9.18EE450 pKa = 3.91IYY452 pKa = 10.8NGARR456 pKa = 11.84LSEE459 pKa = 4.27VPTTLSANYY468 pKa = 8.57EE469 pKa = 4.28HH470 pKa = 6.77GWKK473 pKa = 8.29YY474 pKa = 9.73TSIKK478 pKa = 10.52DD479 pKa = 3.63LASSGIYY486 pKa = 10.09SQDD489 pKa = 4.34DD490 pKa = 3.09IDD492 pKa = 3.68KK493 pKa = 10.07LRR495 pKa = 11.84RR496 pKa = 11.84YY497 pKa = 9.94IMAPNEE503 pKa = 3.7RR504 pKa = 11.84TAITNTLSDD513 pKa = 4.17VINGNIEE520 pKa = 4.04LSPDD524 pKa = 3.52LKK526 pKa = 9.93VTNLIGPQTLEE537 pKa = 3.94SLASKK542 pKa = 10.25LSSKK546 pKa = 9.11IWSMFTLFGHH556 pKa = 6.2VSSGVIGIYY565 pKa = 9.62MLARR569 pKa = 11.84LIKK572 pKa = 10.46LIVDD576 pKa = 4.2TAVHH580 pKa = 5.78GTILYY585 pKa = 10.05NLYY588 pKa = 9.99GWSICLIGGIWDD600 pKa = 4.39SITHH604 pKa = 4.87YY605 pKa = 10.65LIYY608 pKa = 10.33RR609 pKa = 11.84KK610 pKa = 9.39RR611 pKa = 11.84HH612 pKa = 4.23EE613 pKa = 4.41TKK615 pKa = 10.67KK616 pKa = 10.95GDD618 pKa = 3.41TTMEE622 pKa = 4.19TEE624 pKa = 4.17SLEE627 pKa = 4.37VSAKK631 pKa = 10.59DD632 pKa = 4.04YY633 pKa = 11.02ISEE636 pKa = 4.27APLKK640 pKa = 10.31NIHH643 pKa = 6.68INQQRR648 pKa = 11.84VTYY651 pKa = 9.31PNLEE655 pKa = 3.76RR656 pKa = 11.84FMPP659 pKa = 4.64
MM1 pKa = 7.73LILEE5 pKa = 4.58SNMVNYY11 pKa = 9.2MIKK14 pKa = 10.59LLFLPTCYY22 pKa = 10.59GFLAFDD28 pKa = 4.41CSNPNINYY36 pKa = 8.29TLVNTRR42 pKa = 11.84EE43 pKa = 3.98VEE45 pKa = 3.98EE46 pKa = 5.25CIISTPEE53 pKa = 3.44IVEE56 pKa = 4.21EE57 pKa = 4.11KK58 pKa = 11.07VSIQLLQLNSIAYY71 pKa = 9.1AKK73 pKa = 10.42YY74 pKa = 9.03VSCKK78 pKa = 9.65VSMNRR83 pKa = 11.84IISYY87 pKa = 10.55CGMNSHH93 pKa = 6.34VSMVHH98 pKa = 5.58NGYY101 pKa = 11.1ASFIVPITNTEE112 pKa = 3.75CWNIHH117 pKa = 5.22RR118 pKa = 11.84DD119 pKa = 3.34RR120 pKa = 11.84RR121 pKa = 11.84YY122 pKa = 9.72ILPNLSVVSNLKK134 pKa = 10.02PNSTIEE140 pKa = 4.04VSSVLSGKK148 pKa = 10.02LFSDD152 pKa = 4.94GKK154 pKa = 11.04CEE156 pKa = 4.04GGSYY160 pKa = 9.4NDD162 pKa = 5.25KK163 pKa = 9.92YY164 pKa = 11.3GSWEE168 pKa = 4.11SVIVQAIYY176 pKa = 10.35KK177 pKa = 9.27IQLTEE182 pKa = 4.61GYY184 pKa = 7.35TQYY187 pKa = 11.55DD188 pKa = 3.38RR189 pKa = 11.84DD190 pKa = 3.68TNYY193 pKa = 9.75IHH195 pKa = 7.21LPYY198 pKa = 10.27GINCIYY204 pKa = 10.95DD205 pKa = 4.3RR206 pKa = 11.84IDD208 pKa = 3.79CVHH211 pKa = 7.42PDD213 pKa = 2.9IGMTTWDD220 pKa = 3.46VFPTHH225 pKa = 6.55EE226 pKa = 4.79CDD228 pKa = 4.28DD229 pKa = 3.8NRR231 pKa = 11.84HH232 pKa = 5.64LVLFDD237 pKa = 3.65GFASKK242 pKa = 7.73TTTQSLTGEE251 pKa = 3.99NMKK254 pKa = 10.33VVTYY258 pKa = 9.0MVTSSTRR265 pKa = 11.84AFALSVTTRR274 pKa = 11.84YY275 pKa = 9.63DD276 pKa = 3.22KK277 pKa = 11.34CSMEE281 pKa = 4.3VWRR284 pKa = 11.84TEE286 pKa = 3.86HH287 pKa = 6.71PRR289 pKa = 11.84LLILVVNQQWDD300 pKa = 4.03KK301 pKa = 11.13IKK303 pKa = 11.11SNSIDD308 pKa = 3.34ISSLDD313 pKa = 3.29LMAYY317 pKa = 10.19VNAKK321 pKa = 10.37LIMIQASVASNVEE334 pKa = 4.03SLYY337 pKa = 9.36QTLDD341 pKa = 3.01YY342 pKa = 9.79RR343 pKa = 11.84RR344 pKa = 11.84CMAEE348 pKa = 3.86RR349 pKa = 11.84KK350 pKa = 9.46SLRR353 pKa = 11.84NQLALASLSPQEE365 pKa = 3.87FAYY368 pKa = 10.65LYY370 pKa = 8.9MGKK373 pKa = 9.26PGYY376 pKa = 9.04TSLPMGEE383 pKa = 3.9VSYY386 pKa = 10.52IIQCNPVEE394 pKa = 4.34VKK396 pKa = 10.24LRR398 pKa = 11.84KK399 pKa = 8.07TDD401 pKa = 3.05KK402 pKa = 10.43CYY404 pKa = 10.82HH405 pKa = 6.38EE406 pKa = 5.62LPISLNNQSFYY417 pKa = 10.58MSPKK421 pKa = 8.62TRR423 pKa = 11.84LIQSQGTEE431 pKa = 4.18VEE433 pKa = 4.86CNHH436 pKa = 6.61LMLPKK441 pKa = 10.33FLLGKK446 pKa = 9.57NWYY449 pKa = 9.18EE450 pKa = 3.91IYY452 pKa = 10.8NGARR456 pKa = 11.84LSEE459 pKa = 4.27VPTTLSANYY468 pKa = 8.57EE469 pKa = 4.28HH470 pKa = 6.77GWKK473 pKa = 8.29YY474 pKa = 9.73TSIKK478 pKa = 10.52DD479 pKa = 3.63LASSGIYY486 pKa = 10.09SQDD489 pKa = 4.34DD490 pKa = 3.09IDD492 pKa = 3.68KK493 pKa = 10.07LRR495 pKa = 11.84RR496 pKa = 11.84YY497 pKa = 9.94IMAPNEE503 pKa = 3.7RR504 pKa = 11.84TAITNTLSDD513 pKa = 4.17VINGNIEE520 pKa = 4.04LSPDD524 pKa = 3.52LKK526 pKa = 9.93VTNLIGPQTLEE537 pKa = 3.94SLASKK542 pKa = 10.25LSSKK546 pKa = 9.11IWSMFTLFGHH556 pKa = 6.2VSSGVIGIYY565 pKa = 9.62MLARR569 pKa = 11.84LIKK572 pKa = 10.46LIVDD576 pKa = 4.2TAVHH580 pKa = 5.78GTILYY585 pKa = 10.05NLYY588 pKa = 9.99GWSICLIGGIWDD600 pKa = 4.39SITHH604 pKa = 4.87YY605 pKa = 10.65LIYY608 pKa = 10.33RR609 pKa = 11.84KK610 pKa = 9.39RR611 pKa = 11.84HH612 pKa = 4.23EE613 pKa = 4.41TKK615 pKa = 10.67KK616 pKa = 10.95GDD618 pKa = 3.41TTMEE622 pKa = 4.19TEE624 pKa = 4.17SLEE627 pKa = 4.37VSAKK631 pKa = 10.59DD632 pKa = 4.04YY633 pKa = 11.02ISEE636 pKa = 4.27APLKK640 pKa = 10.31NIHH643 pKa = 6.68INQQRR648 pKa = 11.84VTYY651 pKa = 9.31PNLEE655 pKa = 3.76RR656 pKa = 11.84FMPP659 pKa = 4.64
Molecular weight: 75.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KRA0|A0A0B5KRA0_9VIRU Large structural protein OS=Lishi spider virus 1 OX=1608057 GN=L PE=4 SV=1
MM1 pKa = 7.61ANDD4 pKa = 3.11NVMRR8 pKa = 11.84LLGRR12 pKa = 11.84ALNEE16 pKa = 3.91PPIDD20 pKa = 3.75EE21 pKa = 4.47PVGLRR26 pKa = 11.84GPCMNTAFFSKK37 pKa = 9.9WLRR40 pKa = 11.84QDD42 pKa = 3.93IIFQKK47 pKa = 10.65VGVRR51 pKa = 11.84SIDD54 pKa = 3.38VAPFQGDD61 pKa = 3.13FDD63 pKa = 6.0DD64 pKa = 5.06NLVPTMIHH72 pKa = 6.96LYY74 pKa = 10.56SIGKK78 pKa = 8.46FLHH81 pKa = 6.22YY82 pKa = 10.03KK83 pKa = 8.7PQSTVSRR90 pKa = 11.84AAVLAITCAEE100 pKa = 4.09TLGPSIGTDD109 pKa = 2.55RR110 pKa = 11.84LMTQHH115 pKa = 7.05EE116 pKa = 4.38ARR118 pKa = 11.84QSLCHH123 pKa = 6.04AVCALRR129 pKa = 11.84NSGLLATFTSATAQTYY145 pKa = 9.15GALVGNDD152 pKa = 4.47RR153 pKa = 11.84PAQWTDD159 pKa = 3.15TDD161 pKa = 3.54WDD163 pKa = 4.03STDD166 pKa = 3.49VEE168 pKa = 4.67ANCRR172 pKa = 11.84VLRR175 pKa = 11.84HH176 pKa = 4.93MALRR180 pKa = 11.84AEE182 pKa = 4.32AAYY185 pKa = 8.54TLKK188 pKa = 10.67GSSLIVHH195 pKa = 6.5MLVAIIKK202 pKa = 9.78RR203 pKa = 11.84GTMSDD208 pKa = 3.22AFVDD212 pKa = 4.93KK213 pKa = 10.23IQQGIQTDD221 pKa = 4.16LSLPIRR227 pKa = 11.84ISAEE231 pKa = 3.76TCRR234 pKa = 11.84IFYY237 pKa = 10.58SSYY240 pKa = 10.36CSEE243 pKa = 4.31LNDD246 pKa = 3.37VGMSFLLSHH255 pKa = 7.75LDD257 pKa = 3.5RR258 pKa = 11.84LIPTHH263 pKa = 6.65ALRR266 pKa = 11.84LRR268 pKa = 11.84LTIQQASGSGLTALVLLGRR287 pKa = 11.84AFRR290 pKa = 11.84MYY292 pKa = 11.23EE293 pKa = 3.83NFNWPLVAKK302 pKa = 8.9WYY304 pKa = 9.05PDD306 pKa = 2.64EE307 pKa = 4.2WANFTAAVDD316 pKa = 4.0SVGNNLMYY324 pKa = 10.53GYY326 pKa = 10.36RR327 pKa = 11.84RR328 pKa = 11.84DD329 pKa = 3.35IGPAKK334 pKa = 8.97STKK337 pKa = 9.29FRR339 pKa = 11.84SLTFIAKK346 pKa = 9.62EE347 pKa = 3.46LLIRR351 pKa = 11.84VGGEE355 pKa = 3.68TSLRR359 pKa = 11.84NYY361 pKa = 10.61AGFATRR367 pKa = 11.84VKK369 pKa = 10.82NHH371 pKa = 6.98DD372 pKa = 3.52AVINLINTYY381 pKa = 8.24VDD383 pKa = 4.67RR384 pKa = 11.84ITAAQAEE391 pKa = 4.8VEE393 pKa = 4.35VGDD396 pKa = 4.49AEE398 pKa = 4.55SEE400 pKa = 4.13AVNQFRR406 pKa = 11.84QTVVEE411 pKa = 4.04HH412 pKa = 6.42ANVYY416 pKa = 9.62GG417 pKa = 3.59
MM1 pKa = 7.61ANDD4 pKa = 3.11NVMRR8 pKa = 11.84LLGRR12 pKa = 11.84ALNEE16 pKa = 3.91PPIDD20 pKa = 3.75EE21 pKa = 4.47PVGLRR26 pKa = 11.84GPCMNTAFFSKK37 pKa = 9.9WLRR40 pKa = 11.84QDD42 pKa = 3.93IIFQKK47 pKa = 10.65VGVRR51 pKa = 11.84SIDD54 pKa = 3.38VAPFQGDD61 pKa = 3.13FDD63 pKa = 6.0DD64 pKa = 5.06NLVPTMIHH72 pKa = 6.96LYY74 pKa = 10.56SIGKK78 pKa = 8.46FLHH81 pKa = 6.22YY82 pKa = 10.03KK83 pKa = 8.7PQSTVSRR90 pKa = 11.84AAVLAITCAEE100 pKa = 4.09TLGPSIGTDD109 pKa = 2.55RR110 pKa = 11.84LMTQHH115 pKa = 7.05EE116 pKa = 4.38ARR118 pKa = 11.84QSLCHH123 pKa = 6.04AVCALRR129 pKa = 11.84NSGLLATFTSATAQTYY145 pKa = 9.15GALVGNDD152 pKa = 4.47RR153 pKa = 11.84PAQWTDD159 pKa = 3.15TDD161 pKa = 3.54WDD163 pKa = 4.03STDD166 pKa = 3.49VEE168 pKa = 4.67ANCRR172 pKa = 11.84VLRR175 pKa = 11.84HH176 pKa = 4.93MALRR180 pKa = 11.84AEE182 pKa = 4.32AAYY185 pKa = 8.54TLKK188 pKa = 10.67GSSLIVHH195 pKa = 6.5MLVAIIKK202 pKa = 9.78RR203 pKa = 11.84GTMSDD208 pKa = 3.22AFVDD212 pKa = 4.93KK213 pKa = 10.23IQQGIQTDD221 pKa = 4.16LSLPIRR227 pKa = 11.84ISAEE231 pKa = 3.76TCRR234 pKa = 11.84IFYY237 pKa = 10.58SSYY240 pKa = 10.36CSEE243 pKa = 4.31LNDD246 pKa = 3.37VGMSFLLSHH255 pKa = 7.75LDD257 pKa = 3.5RR258 pKa = 11.84LIPTHH263 pKa = 6.65ALRR266 pKa = 11.84LRR268 pKa = 11.84LTIQQASGSGLTALVLLGRR287 pKa = 11.84AFRR290 pKa = 11.84MYY292 pKa = 11.23EE293 pKa = 3.83NFNWPLVAKK302 pKa = 8.9WYY304 pKa = 9.05PDD306 pKa = 2.64EE307 pKa = 4.2WANFTAAVDD316 pKa = 4.0SVGNNLMYY324 pKa = 10.53GYY326 pKa = 10.36RR327 pKa = 11.84RR328 pKa = 11.84DD329 pKa = 3.35IGPAKK334 pKa = 8.97STKK337 pKa = 9.29FRR339 pKa = 11.84SLTFIAKK346 pKa = 9.62EE347 pKa = 3.46LLIRR351 pKa = 11.84VGGEE355 pKa = 3.68TSLRR359 pKa = 11.84NYY361 pKa = 10.61AGFATRR367 pKa = 11.84VKK369 pKa = 10.82NHH371 pKa = 6.98DD372 pKa = 3.52AVINLINTYY381 pKa = 8.24VDD383 pKa = 4.67RR384 pKa = 11.84ITAAQAEE391 pKa = 4.8VEE393 pKa = 4.35VGDD396 pKa = 4.49AEE398 pKa = 4.55SEE400 pKa = 4.13AVNQFRR406 pKa = 11.84QTVVEE411 pKa = 4.04HH412 pKa = 6.42ANVYY416 pKa = 9.62GG417 pKa = 3.59
Molecular weight: 46.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3256 |
417 |
2180 |
1085.3 |
124.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.129 ± 1.14 | 2.119 ± 0.113 |
4.668 ± 0.238 | 6.081 ± 0.635 |
4.146 ± 0.687 | 4.361 ± 0.535 |
2.887 ± 0.249 | 7.033 ± 0.47 |
5.006 ± 0.483 | 10.565 ± 0.331 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.488 ± 0.328 | 5.283 ± 0.302 |
4.115 ± 0.309 | 3.194 ± 0.141 |
5.59 ± 0.481 | 8.016 ± 0.504 |
6.91 ± 0.026 | 6.081 ± 0.339 |
1.413 ± 0.041 | 4.914 ± 0.44 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
