
Penicillium roqueforti ssRNA mycovirus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; Fusariviridae
Average proteome isoelectric point is 8.04
Get precalculated fractions of proteins
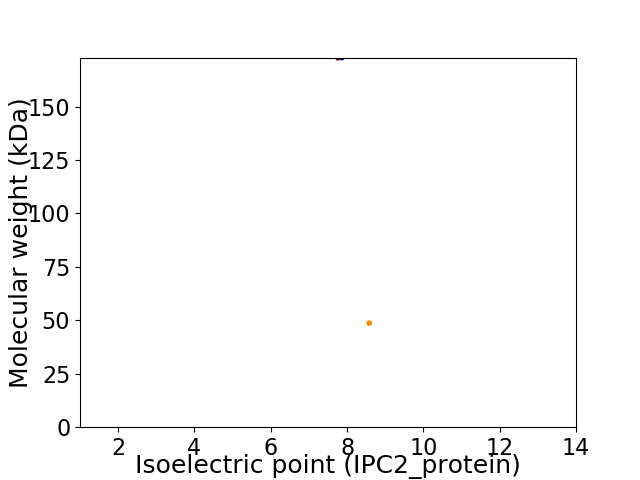
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A076KXD4|A0A076KXD4_9VIRU RNA-directed RNA polymerase OS=Penicillium roqueforti ssRNA mycovirus 1 OX=1532180 PE=4 SV=1
MM1 pKa = 7.78FYY3 pKa = 10.57TFALIMWWSTLHH15 pKa = 6.7ALFGFLCCFGPVYY28 pKa = 10.63LVLALFAVASPVSVLDD44 pKa = 3.72SLLLCVTFPWVVVFIFRR61 pKa = 11.84SWRR64 pKa = 11.84SRR66 pKa = 11.84LDD68 pKa = 3.26QEE70 pKa = 4.73VILINSVGKK79 pKa = 10.36DD80 pKa = 2.97EE81 pKa = 5.6ALWHH85 pKa = 6.74DD86 pKa = 4.05LDD88 pKa = 4.46LFEE91 pKa = 5.74GKK93 pKa = 9.88VAFRR97 pKa = 11.84AVGMQGWDD105 pKa = 3.5LLSPYY110 pKa = 10.03IIRR113 pKa = 11.84VLGDD117 pKa = 3.18VGLSEE122 pKa = 4.71VPLEE126 pKa = 4.21GDD128 pKa = 4.72LRR130 pKa = 11.84DD131 pKa = 4.04GLYY134 pKa = 10.34KK135 pKa = 10.71ASSRR139 pKa = 11.84WWSNCVLSIHH149 pKa = 6.58AVASHH154 pKa = 7.06RR155 pKa = 11.84FTPWFYY161 pKa = 11.38LSFITFSFIIRR172 pKa = 11.84IPVFIAKK179 pKa = 10.0KK180 pKa = 10.17GLALLRR186 pKa = 11.84FWALGALFIWHH197 pKa = 6.63MPDD200 pKa = 2.74EE201 pKa = 4.78LAAKK205 pKa = 10.32LYY207 pKa = 6.02TTLVEE212 pKa = 4.38VSSLFYY218 pKa = 10.61DD219 pKa = 3.93GKK221 pKa = 9.95FVEE224 pKa = 4.6WVGWQLTRR232 pKa = 11.84LSVRR236 pKa = 11.84VTVVLLDD243 pKa = 3.48AQFIGVKK250 pKa = 6.93WTTRR254 pKa = 11.84KK255 pKa = 9.98GFRR258 pKa = 11.84SDD260 pKa = 2.98SEE262 pKa = 3.91ARR264 pKa = 11.84KK265 pKa = 9.2FSSVLRR271 pKa = 11.84EE272 pKa = 3.63GLMQFSIFVTDD283 pKa = 4.63LGLPHH288 pKa = 7.13YY289 pKa = 10.14IRR291 pKa = 11.84GSRR294 pKa = 11.84EE295 pKa = 3.5PTRR298 pKa = 11.84QGVQEE303 pKa = 4.16SYY305 pKa = 11.39EE306 pKa = 3.96LLKK309 pKa = 10.95EE310 pKa = 4.15LGWPVNVGLQDD321 pKa = 3.79PDD323 pKa = 3.85LSVVPEE329 pKa = 4.27KK330 pKa = 10.19WSSWVISGTDD340 pKa = 3.14WQQGINNMKK349 pKa = 8.62THH351 pKa = 6.51VDD353 pKa = 3.13HH354 pKa = 7.64DD355 pKa = 4.33LDD357 pKa = 3.72KK358 pKa = 11.42LRR360 pKa = 11.84LHH362 pKa = 6.87AIEE365 pKa = 3.81YY366 pKa = 10.34RR367 pKa = 11.84RR368 pKa = 11.84TEE370 pKa = 3.99EE371 pKa = 3.96YY372 pKa = 11.07ASVEE376 pKa = 4.14NEE378 pKa = 3.79LEE380 pKa = 3.82ATSRR384 pKa = 11.84YY385 pKa = 8.82FRR387 pKa = 11.84SPRR390 pKa = 11.84YY391 pKa = 9.63DD392 pKa = 3.67YY393 pKa = 11.11PDD395 pKa = 3.81LALEE399 pKa = 4.8DD400 pKa = 3.7VWFMIKK406 pKa = 9.4DD407 pKa = 3.68TFVHH411 pKa = 6.64SRR413 pKa = 11.84LTPFNHH419 pKa = 7.74IIRR422 pKa = 11.84MWEE425 pKa = 3.69KK426 pKa = 10.61KK427 pKa = 9.95YY428 pKa = 11.19GLGAFFRR435 pKa = 11.84RR436 pKa = 11.84PGSKK440 pKa = 10.47AKK442 pKa = 9.48MRR444 pKa = 11.84RR445 pKa = 11.84SDD447 pKa = 4.23FISSIGGYY455 pKa = 10.78APFKK459 pKa = 10.02RR460 pKa = 11.84LWRR463 pKa = 11.84ATFEE467 pKa = 4.35VATSIVPVSAVSVKK481 pKa = 10.89NEE483 pKa = 3.81ALPPNKK489 pKa = 9.07WAEE492 pKa = 4.02NKK494 pKa = 9.25VRR496 pKa = 11.84SIIGSPIAHH505 pKa = 6.75YY506 pKa = 10.44ILSTIWNYY514 pKa = 10.38EE515 pKa = 3.86PNHH518 pKa = 5.75RR519 pKa = 11.84FAWTTTPTKK528 pKa = 10.09IGMPLNGYY536 pKa = 8.29WLADD540 pKa = 3.68LYY542 pKa = 10.96HH543 pKa = 6.22RR544 pKa = 11.84HH545 pKa = 6.01SRR547 pKa = 11.84CQHH550 pKa = 5.42HH551 pKa = 6.64VAGDD555 pKa = 3.46MSAFDD560 pKa = 3.68STLSGEE566 pKa = 4.47VVKK569 pKa = 10.37MIAAVRR575 pKa = 11.84KK576 pKa = 10.1KK577 pKa = 10.47GFEE580 pKa = 3.84SHH582 pKa = 7.0KK583 pKa = 10.85DD584 pKa = 2.98VDD586 pKa = 4.92RR587 pKa = 11.84ICDD590 pKa = 4.56LIDD593 pKa = 3.25VSYY596 pKa = 10.6EE597 pKa = 3.95QLGHH601 pKa = 6.0QLLNTTSTGNIYY613 pKa = 10.88GKK615 pKa = 9.19GTGLTTGHH623 pKa = 6.63SSTSMDD629 pKa = 3.14NSLGLLILYY638 pKa = 9.75LMAWKK643 pKa = 10.2QLTGLGARR651 pKa = 11.84EE652 pKa = 3.55FKK654 pKa = 10.78HH655 pKa = 6.49FNEE658 pKa = 4.33LSDD661 pKa = 3.92YY662 pKa = 11.26GDD664 pKa = 3.8DD665 pKa = 5.08HH666 pKa = 7.71ILSYY670 pKa = 11.31LSTKK674 pKa = 8.4PAAWNFRR681 pKa = 11.84NIQKK685 pKa = 9.93VMARR689 pKa = 11.84WGVTNRR695 pKa = 11.84EE696 pKa = 3.99EE697 pKa = 4.65KK698 pKa = 10.09MGSLDD703 pKa = 4.35VIPFLSKK710 pKa = 9.82TSRR713 pKa = 11.84KK714 pKa = 7.15VTNEE718 pKa = 3.46DD719 pKa = 2.83RR720 pKa = 11.84VSFAKK725 pKa = 10.87YY726 pKa = 9.63GVKK729 pKa = 9.96EE730 pKa = 4.11PKK732 pKa = 9.95RR733 pKa = 11.84VVLHH737 pKa = 6.76DD738 pKa = 3.54RR739 pKa = 11.84EE740 pKa = 4.2KK741 pKa = 11.23LVGKK745 pKa = 9.03MVAKK749 pKa = 10.24VKK751 pKa = 10.76NLDD754 pKa = 3.14PVYY757 pKa = 9.97RR758 pKa = 11.84AKK760 pKa = 10.8RR761 pKa = 11.84LISYY765 pKa = 8.33MGLTAHH771 pKa = 7.25HH772 pKa = 7.25EE773 pKa = 4.04DD774 pKa = 4.56LYY776 pKa = 11.65SGISDD781 pKa = 3.44VLARR785 pKa = 11.84SSTLRR790 pKa = 11.84RR791 pKa = 11.84AVRR794 pKa = 11.84KK795 pKa = 9.38EE796 pKa = 3.48GLKK799 pKa = 9.97IPTYY803 pKa = 10.77KK804 pKa = 10.34KK805 pKa = 10.64VLQQWYY811 pKa = 9.15NPSANISHH819 pKa = 7.33DD820 pKa = 4.02VIQEE824 pKa = 4.03EE825 pKa = 4.7LSDD828 pKa = 4.01GQKK831 pKa = 11.13SMVMHH836 pKa = 6.58YY837 pKa = 10.53GQVTIIDD844 pKa = 4.54SISGALAMLPDD855 pKa = 4.16ILNPAIFNFGYY866 pKa = 10.41SRR868 pKa = 11.84LLQLQLRR875 pKa = 11.84SFLEE879 pKa = 4.05WPIEE883 pKa = 3.84FLVMQNGVVSQAEE896 pKa = 4.12LLRR899 pKa = 11.84IFRR902 pKa = 11.84STAYY906 pKa = 9.86EE907 pKa = 3.93AVDD910 pKa = 3.65PTIHH914 pKa = 5.8TMSGASYY921 pKa = 10.61DD922 pKa = 3.69YY923 pKa = 11.2GGLLLRR929 pKa = 11.84HH930 pKa = 6.24WLFCWYY936 pKa = 9.91KK937 pKa = 10.33GRR939 pKa = 11.84SLAPSVLSWFEE950 pKa = 3.92KK951 pKa = 10.77VVGKK955 pKa = 10.93VNLVQFLLNGRR966 pKa = 11.84ANIQLGRR973 pKa = 11.84ANPYY977 pKa = 10.7LLDD980 pKa = 4.02LFVISILNLVPSVPILSPLKK1000 pKa = 10.22SIRR1003 pKa = 11.84LPRR1006 pKa = 11.84VDD1008 pKa = 4.39IFLDD1012 pKa = 3.78MVWQVLVVFVWSGVPPNYY1030 pKa = 10.1KK1031 pKa = 10.13EE1032 pKa = 3.98LTHH1035 pKa = 7.46LARR1038 pKa = 11.84QLQEE1042 pKa = 3.99LKK1044 pKa = 11.05GGVLVTAPTGTGKK1057 pKa = 8.55STTMVKK1063 pKa = 10.28HH1064 pKa = 6.55LEE1066 pKa = 4.02LTLGLFYY1073 pKa = 11.21NKK1075 pKa = 9.67IVIIEE1080 pKa = 4.04PRR1082 pKa = 11.84SALVKK1087 pKa = 10.83SLVPYY1092 pKa = 10.39CKK1094 pKa = 9.12NTLNMSCTGRR1104 pKa = 11.84TMGFDD1109 pKa = 3.58FDD1111 pKa = 4.04RR1112 pKa = 11.84RR1113 pKa = 11.84EE1114 pKa = 4.15KK1115 pKa = 9.97VWYY1118 pKa = 7.48VTPQEE1123 pKa = 4.52AALHH1127 pKa = 5.3WEE1129 pKa = 4.27EE1130 pKa = 4.2TFEE1133 pKa = 4.17EE1134 pKa = 4.41NNLIMIDD1141 pKa = 3.59EE1142 pKa = 4.2AHH1144 pKa = 6.64IEE1146 pKa = 3.98EE1147 pKa = 4.68PFYY1150 pKa = 11.71VFLKK1154 pKa = 10.53EE1155 pKa = 3.96VLKK1158 pKa = 11.16SSGRR1162 pKa = 11.84GHH1164 pKa = 7.32LLVTATPTPSLLDD1177 pKa = 3.32SCVAQVPLLLAQLWTVLSSKK1197 pKa = 10.11DD1198 pKa = 3.27SCMEE1202 pKa = 4.0SDD1204 pKa = 3.78PRR1206 pKa = 11.84TCIKK1210 pKa = 10.36RR1211 pKa = 11.84YY1212 pKa = 9.12EE1213 pKa = 4.24DD1214 pKa = 3.22WVVDD1218 pKa = 3.88QVHH1221 pKa = 6.22NSWTNSKK1228 pKa = 10.7ALVFHH1233 pKa = 6.82PSKK1236 pKa = 11.08DD1237 pKa = 3.38GAARR1241 pKa = 11.84LADD1244 pKa = 4.92RR1245 pKa = 11.84ISRR1248 pKa = 11.84KK1249 pKa = 9.7CSFYY1253 pKa = 11.26NSDD1256 pKa = 3.53VQDD1259 pKa = 3.66GSGQVILSTNVTDD1272 pKa = 6.0AGLTLPNVDD1281 pKa = 4.61LVLTSEE1287 pKa = 5.2LDD1289 pKa = 3.63NVMDD1293 pKa = 4.84PAAGTVVLGRR1303 pKa = 11.84LSLQTTYY1310 pKa = 9.65QRR1312 pKa = 11.84RR1313 pKa = 11.84GRR1315 pKa = 11.84TGRR1318 pKa = 11.84TSNGQFKK1325 pKa = 10.96LFACPNANVKK1335 pKa = 10.2QIAEE1339 pKa = 3.78IALNEE1344 pKa = 3.99KK1345 pKa = 10.59SLVSEE1350 pKa = 4.13WLSLGLSPWLIHH1362 pKa = 6.43TIDD1365 pKa = 3.83SKK1367 pKa = 11.75RR1368 pKa = 11.84MFSFLGMDD1376 pKa = 4.16SEE1378 pKa = 5.44RR1379 pKa = 11.84VDD1381 pKa = 3.54PDD1383 pKa = 3.68LLRR1386 pKa = 11.84EE1387 pKa = 4.23TLNSLQLFSNNLEE1400 pKa = 3.89FLRR1403 pKa = 11.84ASRR1406 pKa = 11.84AAQEE1410 pKa = 4.03AQKK1413 pKa = 11.25AEE1415 pKa = 4.13DD1416 pKa = 4.02PNKK1419 pKa = 10.03FVYY1422 pKa = 10.09DD1423 pKa = 3.87YY1424 pKa = 10.13SASGIIRR1431 pKa = 11.84DD1432 pKa = 3.49SSTIDD1437 pKa = 3.36FSEE1440 pKa = 4.1ISRR1443 pKa = 11.84IALRR1447 pKa = 11.84VAALGPSLKK1456 pKa = 10.47RR1457 pKa = 11.84GTDD1460 pKa = 2.98VDD1462 pKa = 4.3AGQLDD1467 pKa = 3.78SDD1469 pKa = 5.28LFFLDD1474 pKa = 4.0QISGVHH1480 pKa = 5.26TPFKK1484 pKa = 11.25NLMPDD1489 pKa = 3.6LTGLLEE1495 pKa = 4.26KK1496 pKa = 10.08TDD1498 pKa = 3.74YY1499 pKa = 10.88DD1500 pKa = 4.0YY1501 pKa = 12.02KK1502 pKa = 10.62EE1503 pKa = 4.08VQAIKK1508 pKa = 10.02PPKK1511 pKa = 9.24ILHH1514 pKa = 5.81KK1515 pKa = 10.48GCRR1518 pKa = 11.84EE1519 pKa = 3.66PDD1521 pKa = 3.3EE1522 pKa = 4.31PP1523 pKa = 5.59
MM1 pKa = 7.78FYY3 pKa = 10.57TFALIMWWSTLHH15 pKa = 6.7ALFGFLCCFGPVYY28 pKa = 10.63LVLALFAVASPVSVLDD44 pKa = 3.72SLLLCVTFPWVVVFIFRR61 pKa = 11.84SWRR64 pKa = 11.84SRR66 pKa = 11.84LDD68 pKa = 3.26QEE70 pKa = 4.73VILINSVGKK79 pKa = 10.36DD80 pKa = 2.97EE81 pKa = 5.6ALWHH85 pKa = 6.74DD86 pKa = 4.05LDD88 pKa = 4.46LFEE91 pKa = 5.74GKK93 pKa = 9.88VAFRR97 pKa = 11.84AVGMQGWDD105 pKa = 3.5LLSPYY110 pKa = 10.03IIRR113 pKa = 11.84VLGDD117 pKa = 3.18VGLSEE122 pKa = 4.71VPLEE126 pKa = 4.21GDD128 pKa = 4.72LRR130 pKa = 11.84DD131 pKa = 4.04GLYY134 pKa = 10.34KK135 pKa = 10.71ASSRR139 pKa = 11.84WWSNCVLSIHH149 pKa = 6.58AVASHH154 pKa = 7.06RR155 pKa = 11.84FTPWFYY161 pKa = 11.38LSFITFSFIIRR172 pKa = 11.84IPVFIAKK179 pKa = 10.0KK180 pKa = 10.17GLALLRR186 pKa = 11.84FWALGALFIWHH197 pKa = 6.63MPDD200 pKa = 2.74EE201 pKa = 4.78LAAKK205 pKa = 10.32LYY207 pKa = 6.02TTLVEE212 pKa = 4.38VSSLFYY218 pKa = 10.61DD219 pKa = 3.93GKK221 pKa = 9.95FVEE224 pKa = 4.6WVGWQLTRR232 pKa = 11.84LSVRR236 pKa = 11.84VTVVLLDD243 pKa = 3.48AQFIGVKK250 pKa = 6.93WTTRR254 pKa = 11.84KK255 pKa = 9.98GFRR258 pKa = 11.84SDD260 pKa = 2.98SEE262 pKa = 3.91ARR264 pKa = 11.84KK265 pKa = 9.2FSSVLRR271 pKa = 11.84EE272 pKa = 3.63GLMQFSIFVTDD283 pKa = 4.63LGLPHH288 pKa = 7.13YY289 pKa = 10.14IRR291 pKa = 11.84GSRR294 pKa = 11.84EE295 pKa = 3.5PTRR298 pKa = 11.84QGVQEE303 pKa = 4.16SYY305 pKa = 11.39EE306 pKa = 3.96LLKK309 pKa = 10.95EE310 pKa = 4.15LGWPVNVGLQDD321 pKa = 3.79PDD323 pKa = 3.85LSVVPEE329 pKa = 4.27KK330 pKa = 10.19WSSWVISGTDD340 pKa = 3.14WQQGINNMKK349 pKa = 8.62THH351 pKa = 6.51VDD353 pKa = 3.13HH354 pKa = 7.64DD355 pKa = 4.33LDD357 pKa = 3.72KK358 pKa = 11.42LRR360 pKa = 11.84LHH362 pKa = 6.87AIEE365 pKa = 3.81YY366 pKa = 10.34RR367 pKa = 11.84RR368 pKa = 11.84TEE370 pKa = 3.99EE371 pKa = 3.96YY372 pKa = 11.07ASVEE376 pKa = 4.14NEE378 pKa = 3.79LEE380 pKa = 3.82ATSRR384 pKa = 11.84YY385 pKa = 8.82FRR387 pKa = 11.84SPRR390 pKa = 11.84YY391 pKa = 9.63DD392 pKa = 3.67YY393 pKa = 11.11PDD395 pKa = 3.81LALEE399 pKa = 4.8DD400 pKa = 3.7VWFMIKK406 pKa = 9.4DD407 pKa = 3.68TFVHH411 pKa = 6.64SRR413 pKa = 11.84LTPFNHH419 pKa = 7.74IIRR422 pKa = 11.84MWEE425 pKa = 3.69KK426 pKa = 10.61KK427 pKa = 9.95YY428 pKa = 11.19GLGAFFRR435 pKa = 11.84RR436 pKa = 11.84PGSKK440 pKa = 10.47AKK442 pKa = 9.48MRR444 pKa = 11.84RR445 pKa = 11.84SDD447 pKa = 4.23FISSIGGYY455 pKa = 10.78APFKK459 pKa = 10.02RR460 pKa = 11.84LWRR463 pKa = 11.84ATFEE467 pKa = 4.35VATSIVPVSAVSVKK481 pKa = 10.89NEE483 pKa = 3.81ALPPNKK489 pKa = 9.07WAEE492 pKa = 4.02NKK494 pKa = 9.25VRR496 pKa = 11.84SIIGSPIAHH505 pKa = 6.75YY506 pKa = 10.44ILSTIWNYY514 pKa = 10.38EE515 pKa = 3.86PNHH518 pKa = 5.75RR519 pKa = 11.84FAWTTTPTKK528 pKa = 10.09IGMPLNGYY536 pKa = 8.29WLADD540 pKa = 3.68LYY542 pKa = 10.96HH543 pKa = 6.22RR544 pKa = 11.84HH545 pKa = 6.01SRR547 pKa = 11.84CQHH550 pKa = 5.42HH551 pKa = 6.64VAGDD555 pKa = 3.46MSAFDD560 pKa = 3.68STLSGEE566 pKa = 4.47VVKK569 pKa = 10.37MIAAVRR575 pKa = 11.84KK576 pKa = 10.1KK577 pKa = 10.47GFEE580 pKa = 3.84SHH582 pKa = 7.0KK583 pKa = 10.85DD584 pKa = 2.98VDD586 pKa = 4.92RR587 pKa = 11.84ICDD590 pKa = 4.56LIDD593 pKa = 3.25VSYY596 pKa = 10.6EE597 pKa = 3.95QLGHH601 pKa = 6.0QLLNTTSTGNIYY613 pKa = 10.88GKK615 pKa = 9.19GTGLTTGHH623 pKa = 6.63SSTSMDD629 pKa = 3.14NSLGLLILYY638 pKa = 9.75LMAWKK643 pKa = 10.2QLTGLGARR651 pKa = 11.84EE652 pKa = 3.55FKK654 pKa = 10.78HH655 pKa = 6.49FNEE658 pKa = 4.33LSDD661 pKa = 3.92YY662 pKa = 11.26GDD664 pKa = 3.8DD665 pKa = 5.08HH666 pKa = 7.71ILSYY670 pKa = 11.31LSTKK674 pKa = 8.4PAAWNFRR681 pKa = 11.84NIQKK685 pKa = 9.93VMARR689 pKa = 11.84WGVTNRR695 pKa = 11.84EE696 pKa = 3.99EE697 pKa = 4.65KK698 pKa = 10.09MGSLDD703 pKa = 4.35VIPFLSKK710 pKa = 9.82TSRR713 pKa = 11.84KK714 pKa = 7.15VTNEE718 pKa = 3.46DD719 pKa = 2.83RR720 pKa = 11.84VSFAKK725 pKa = 10.87YY726 pKa = 9.63GVKK729 pKa = 9.96EE730 pKa = 4.11PKK732 pKa = 9.95RR733 pKa = 11.84VVLHH737 pKa = 6.76DD738 pKa = 3.54RR739 pKa = 11.84EE740 pKa = 4.2KK741 pKa = 11.23LVGKK745 pKa = 9.03MVAKK749 pKa = 10.24VKK751 pKa = 10.76NLDD754 pKa = 3.14PVYY757 pKa = 9.97RR758 pKa = 11.84AKK760 pKa = 10.8RR761 pKa = 11.84LISYY765 pKa = 8.33MGLTAHH771 pKa = 7.25HH772 pKa = 7.25EE773 pKa = 4.04DD774 pKa = 4.56LYY776 pKa = 11.65SGISDD781 pKa = 3.44VLARR785 pKa = 11.84SSTLRR790 pKa = 11.84RR791 pKa = 11.84AVRR794 pKa = 11.84KK795 pKa = 9.38EE796 pKa = 3.48GLKK799 pKa = 9.97IPTYY803 pKa = 10.77KK804 pKa = 10.34KK805 pKa = 10.64VLQQWYY811 pKa = 9.15NPSANISHH819 pKa = 7.33DD820 pKa = 4.02VIQEE824 pKa = 4.03EE825 pKa = 4.7LSDD828 pKa = 4.01GQKK831 pKa = 11.13SMVMHH836 pKa = 6.58YY837 pKa = 10.53GQVTIIDD844 pKa = 4.54SISGALAMLPDD855 pKa = 4.16ILNPAIFNFGYY866 pKa = 10.41SRR868 pKa = 11.84LLQLQLRR875 pKa = 11.84SFLEE879 pKa = 4.05WPIEE883 pKa = 3.84FLVMQNGVVSQAEE896 pKa = 4.12LLRR899 pKa = 11.84IFRR902 pKa = 11.84STAYY906 pKa = 9.86EE907 pKa = 3.93AVDD910 pKa = 3.65PTIHH914 pKa = 5.8TMSGASYY921 pKa = 10.61DD922 pKa = 3.69YY923 pKa = 11.2GGLLLRR929 pKa = 11.84HH930 pKa = 6.24WLFCWYY936 pKa = 9.91KK937 pKa = 10.33GRR939 pKa = 11.84SLAPSVLSWFEE950 pKa = 3.92KK951 pKa = 10.77VVGKK955 pKa = 10.93VNLVQFLLNGRR966 pKa = 11.84ANIQLGRR973 pKa = 11.84ANPYY977 pKa = 10.7LLDD980 pKa = 4.02LFVISILNLVPSVPILSPLKK1000 pKa = 10.22SIRR1003 pKa = 11.84LPRR1006 pKa = 11.84VDD1008 pKa = 4.39IFLDD1012 pKa = 3.78MVWQVLVVFVWSGVPPNYY1030 pKa = 10.1KK1031 pKa = 10.13EE1032 pKa = 3.98LTHH1035 pKa = 7.46LARR1038 pKa = 11.84QLQEE1042 pKa = 3.99LKK1044 pKa = 11.05GGVLVTAPTGTGKK1057 pKa = 8.55STTMVKK1063 pKa = 10.28HH1064 pKa = 6.55LEE1066 pKa = 4.02LTLGLFYY1073 pKa = 11.21NKK1075 pKa = 9.67IVIIEE1080 pKa = 4.04PRR1082 pKa = 11.84SALVKK1087 pKa = 10.83SLVPYY1092 pKa = 10.39CKK1094 pKa = 9.12NTLNMSCTGRR1104 pKa = 11.84TMGFDD1109 pKa = 3.58FDD1111 pKa = 4.04RR1112 pKa = 11.84RR1113 pKa = 11.84EE1114 pKa = 4.15KK1115 pKa = 9.97VWYY1118 pKa = 7.48VTPQEE1123 pKa = 4.52AALHH1127 pKa = 5.3WEE1129 pKa = 4.27EE1130 pKa = 4.2TFEE1133 pKa = 4.17EE1134 pKa = 4.41NNLIMIDD1141 pKa = 3.59EE1142 pKa = 4.2AHH1144 pKa = 6.64IEE1146 pKa = 3.98EE1147 pKa = 4.68PFYY1150 pKa = 11.71VFLKK1154 pKa = 10.53EE1155 pKa = 3.96VLKK1158 pKa = 11.16SSGRR1162 pKa = 11.84GHH1164 pKa = 7.32LLVTATPTPSLLDD1177 pKa = 3.32SCVAQVPLLLAQLWTVLSSKK1197 pKa = 10.11DD1198 pKa = 3.27SCMEE1202 pKa = 4.0SDD1204 pKa = 3.78PRR1206 pKa = 11.84TCIKK1210 pKa = 10.36RR1211 pKa = 11.84YY1212 pKa = 9.12EE1213 pKa = 4.24DD1214 pKa = 3.22WVVDD1218 pKa = 3.88QVHH1221 pKa = 6.22NSWTNSKK1228 pKa = 10.7ALVFHH1233 pKa = 6.82PSKK1236 pKa = 11.08DD1237 pKa = 3.38GAARR1241 pKa = 11.84LADD1244 pKa = 4.92RR1245 pKa = 11.84ISRR1248 pKa = 11.84KK1249 pKa = 9.7CSFYY1253 pKa = 11.26NSDD1256 pKa = 3.53VQDD1259 pKa = 3.66GSGQVILSTNVTDD1272 pKa = 6.0AGLTLPNVDD1281 pKa = 4.61LVLTSEE1287 pKa = 5.2LDD1289 pKa = 3.63NVMDD1293 pKa = 4.84PAAGTVVLGRR1303 pKa = 11.84LSLQTTYY1310 pKa = 9.65QRR1312 pKa = 11.84RR1313 pKa = 11.84GRR1315 pKa = 11.84TGRR1318 pKa = 11.84TSNGQFKK1325 pKa = 10.96LFACPNANVKK1335 pKa = 10.2QIAEE1339 pKa = 3.78IALNEE1344 pKa = 3.99KK1345 pKa = 10.59SLVSEE1350 pKa = 4.13WLSLGLSPWLIHH1362 pKa = 6.43TIDD1365 pKa = 3.83SKK1367 pKa = 11.75RR1368 pKa = 11.84MFSFLGMDD1376 pKa = 4.16SEE1378 pKa = 5.44RR1379 pKa = 11.84VDD1381 pKa = 3.54PDD1383 pKa = 3.68LLRR1386 pKa = 11.84EE1387 pKa = 4.23TLNSLQLFSNNLEE1400 pKa = 3.89FLRR1403 pKa = 11.84ASRR1406 pKa = 11.84AAQEE1410 pKa = 4.03AQKK1413 pKa = 11.25AEE1415 pKa = 4.13DD1416 pKa = 4.02PNKK1419 pKa = 10.03FVYY1422 pKa = 10.09DD1423 pKa = 3.87YY1424 pKa = 10.13SASGIIRR1431 pKa = 11.84DD1432 pKa = 3.49SSTIDD1437 pKa = 3.36FSEE1440 pKa = 4.1ISRR1443 pKa = 11.84IALRR1447 pKa = 11.84VAALGPSLKK1456 pKa = 10.47RR1457 pKa = 11.84GTDD1460 pKa = 2.98VDD1462 pKa = 4.3AGQLDD1467 pKa = 3.78SDD1469 pKa = 5.28LFFLDD1474 pKa = 4.0QISGVHH1480 pKa = 5.26TPFKK1484 pKa = 11.25NLMPDD1489 pKa = 3.6LTGLLEE1495 pKa = 4.26KK1496 pKa = 10.08TDD1498 pKa = 3.74YY1499 pKa = 10.88DD1500 pKa = 4.0YY1501 pKa = 12.02KK1502 pKa = 10.62EE1503 pKa = 4.08VQAIKK1508 pKa = 10.02PPKK1511 pKa = 9.24ILHH1514 pKa = 5.81KK1515 pKa = 10.48GCRR1518 pKa = 11.84EE1519 pKa = 3.66PDD1521 pKa = 3.3EE1522 pKa = 4.31PP1523 pKa = 5.59
Molecular weight: 172.83 kDa
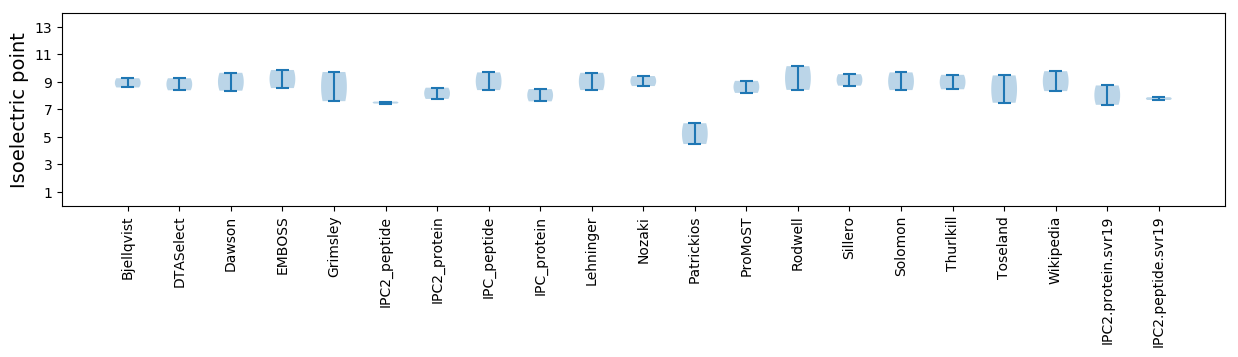
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076KXD4|A0A076KXD4_9VIRU RNA-directed RNA polymerase OS=Penicillium roqueforti ssRNA mycovirus 1 OX=1532180 PE=4 SV=1
MM1 pKa = 7.25TSVNTFNTAEE11 pKa = 4.24TTGALLGEE19 pKa = 4.27QIKK22 pKa = 10.63AIRR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 10.31RR28 pKa = 11.84MEE30 pKa = 3.89NQGFKK35 pKa = 10.91GFWVQAEE42 pKa = 4.31KK43 pKa = 11.05LDD45 pKa = 3.92ALEE48 pKa = 4.85SGLEE52 pKa = 3.85KK53 pKa = 10.64LISGEE58 pKa = 4.21EE59 pKa = 4.06KK60 pKa = 10.15TSLGMSALSSLNEE73 pKa = 3.79EE74 pKa = 4.11LQEE77 pKa = 4.18LRR79 pKa = 11.84RR80 pKa = 11.84EE81 pKa = 3.94NASRR85 pKa = 11.84KK86 pKa = 8.28TILEE90 pKa = 3.67ASKK93 pKa = 10.83FNEE96 pKa = 4.09AKK98 pKa = 10.76LKK100 pKa = 10.96GDD102 pKa = 4.17LEE104 pKa = 4.23QMKK107 pKa = 10.35RR108 pKa = 11.84SADD111 pKa = 3.29AMRR114 pKa = 11.84EE115 pKa = 3.85LSQKK119 pKa = 9.76EE120 pKa = 4.01KK121 pKa = 11.14ADD123 pKa = 3.03IGQTINEE130 pKa = 4.32LKK132 pKa = 10.73EE133 pKa = 4.04EE134 pKa = 4.11LSSARR139 pKa = 11.84KK140 pKa = 9.39SLAASRR146 pKa = 11.84QANRR150 pKa = 11.84EE151 pKa = 4.17SVKK154 pKa = 10.54EE155 pKa = 3.91ATADD159 pKa = 3.53KK160 pKa = 11.03GALQADD166 pKa = 4.89LKK168 pKa = 11.16LKK170 pKa = 10.37ADD172 pKa = 3.53EE173 pKa = 4.19VTRR176 pKa = 11.84LNNLLADD183 pKa = 4.34LNGQVKK189 pKa = 10.64SKK191 pKa = 9.84TKK193 pKa = 10.54AVNDD197 pKa = 4.11AYY199 pKa = 10.89AQVEE203 pKa = 4.39SLKK206 pKa = 10.96ARR208 pKa = 11.84IEE210 pKa = 4.33AEE212 pKa = 3.77SSLQKK217 pKa = 10.21RR218 pKa = 11.84ASGMSYY224 pKa = 11.05SDD226 pKa = 4.35ALKK229 pKa = 10.69QPAPEE234 pKa = 4.94LKK236 pKa = 9.98PLDD239 pKa = 5.18PIPEE243 pKa = 4.08PEE245 pKa = 4.06RR246 pKa = 11.84YY247 pKa = 9.22VLKK250 pKa = 10.64KK251 pKa = 10.46ALKK254 pKa = 9.8SDD256 pKa = 3.53VKK258 pKa = 11.19AKK260 pKa = 10.47LEE262 pKa = 3.91EE263 pKa = 4.01YY264 pKa = 10.55AALKK268 pKa = 10.57RR269 pKa = 11.84EE270 pKa = 4.72AIRR273 pKa = 11.84EE274 pKa = 4.02RR275 pKa = 11.84AHH277 pKa = 7.75KK278 pKa = 10.52LMEE281 pKa = 4.59IANSTDD287 pKa = 2.91PKK289 pKa = 11.24NFVGYY294 pKa = 9.74KK295 pKa = 9.91AYY297 pKa = 10.67ASVLFSKK304 pKa = 10.67IKK306 pKa = 8.95DD307 pKa = 3.74TPVGRR312 pKa = 11.84RR313 pKa = 11.84MSFEE317 pKa = 3.88PAIDD321 pKa = 4.22DD322 pKa = 3.85LWEE325 pKa = 4.51SMAFTSGKK333 pKa = 8.94PLRR336 pKa = 11.84AVKK339 pKa = 10.15AQYY342 pKa = 10.89NEE344 pKa = 4.21LFNTPVEE351 pKa = 4.23EE352 pKa = 4.11QEE354 pKa = 4.12KK355 pKa = 10.15EE356 pKa = 4.13KK357 pKa = 11.04ASSAGSEE364 pKa = 4.29TWWQSTKK371 pKa = 10.43FDD373 pKa = 3.84FWWYY377 pKa = 10.22NATSRR382 pKa = 11.84SWYY385 pKa = 9.98KK386 pKa = 8.56EE387 pKa = 3.44QKK389 pKa = 9.5EE390 pKa = 3.99RR391 pKa = 11.84LVGKK395 pKa = 9.85KK396 pKa = 9.74AQASSWFGRR405 pKa = 11.84LKK407 pKa = 10.72LRR409 pKa = 11.84AEE411 pKa = 4.16SLRR414 pKa = 11.84LYY416 pKa = 9.98IVSFWHH422 pKa = 6.32WGSSVVKK429 pKa = 9.84TDD431 pKa = 2.81
MM1 pKa = 7.25TSVNTFNTAEE11 pKa = 4.24TTGALLGEE19 pKa = 4.27QIKK22 pKa = 10.63AIRR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 10.31RR28 pKa = 11.84MEE30 pKa = 3.89NQGFKK35 pKa = 10.91GFWVQAEE42 pKa = 4.31KK43 pKa = 11.05LDD45 pKa = 3.92ALEE48 pKa = 4.85SGLEE52 pKa = 3.85KK53 pKa = 10.64LISGEE58 pKa = 4.21EE59 pKa = 4.06KK60 pKa = 10.15TSLGMSALSSLNEE73 pKa = 3.79EE74 pKa = 4.11LQEE77 pKa = 4.18LRR79 pKa = 11.84RR80 pKa = 11.84EE81 pKa = 3.94NASRR85 pKa = 11.84KK86 pKa = 8.28TILEE90 pKa = 3.67ASKK93 pKa = 10.83FNEE96 pKa = 4.09AKK98 pKa = 10.76LKK100 pKa = 10.96GDD102 pKa = 4.17LEE104 pKa = 4.23QMKK107 pKa = 10.35RR108 pKa = 11.84SADD111 pKa = 3.29AMRR114 pKa = 11.84EE115 pKa = 3.85LSQKK119 pKa = 9.76EE120 pKa = 4.01KK121 pKa = 11.14ADD123 pKa = 3.03IGQTINEE130 pKa = 4.32LKK132 pKa = 10.73EE133 pKa = 4.04EE134 pKa = 4.11LSSARR139 pKa = 11.84KK140 pKa = 9.39SLAASRR146 pKa = 11.84QANRR150 pKa = 11.84EE151 pKa = 4.17SVKK154 pKa = 10.54EE155 pKa = 3.91ATADD159 pKa = 3.53KK160 pKa = 11.03GALQADD166 pKa = 4.89LKK168 pKa = 11.16LKK170 pKa = 10.37ADD172 pKa = 3.53EE173 pKa = 4.19VTRR176 pKa = 11.84LNNLLADD183 pKa = 4.34LNGQVKK189 pKa = 10.64SKK191 pKa = 9.84TKK193 pKa = 10.54AVNDD197 pKa = 4.11AYY199 pKa = 10.89AQVEE203 pKa = 4.39SLKK206 pKa = 10.96ARR208 pKa = 11.84IEE210 pKa = 4.33AEE212 pKa = 3.77SSLQKK217 pKa = 10.21RR218 pKa = 11.84ASGMSYY224 pKa = 11.05SDD226 pKa = 4.35ALKK229 pKa = 10.69QPAPEE234 pKa = 4.94LKK236 pKa = 9.98PLDD239 pKa = 5.18PIPEE243 pKa = 4.08PEE245 pKa = 4.06RR246 pKa = 11.84YY247 pKa = 9.22VLKK250 pKa = 10.64KK251 pKa = 10.46ALKK254 pKa = 9.8SDD256 pKa = 3.53VKK258 pKa = 11.19AKK260 pKa = 10.47LEE262 pKa = 3.91EE263 pKa = 4.01YY264 pKa = 10.55AALKK268 pKa = 10.57RR269 pKa = 11.84EE270 pKa = 4.72AIRR273 pKa = 11.84EE274 pKa = 4.02RR275 pKa = 11.84AHH277 pKa = 7.75KK278 pKa = 10.52LMEE281 pKa = 4.59IANSTDD287 pKa = 2.91PKK289 pKa = 11.24NFVGYY294 pKa = 9.74KK295 pKa = 9.91AYY297 pKa = 10.67ASVLFSKK304 pKa = 10.67IKK306 pKa = 8.95DD307 pKa = 3.74TPVGRR312 pKa = 11.84RR313 pKa = 11.84MSFEE317 pKa = 3.88PAIDD321 pKa = 4.22DD322 pKa = 3.85LWEE325 pKa = 4.51SMAFTSGKK333 pKa = 8.94PLRR336 pKa = 11.84AVKK339 pKa = 10.15AQYY342 pKa = 10.89NEE344 pKa = 4.21LFNTPVEE351 pKa = 4.23EE352 pKa = 4.11QEE354 pKa = 4.12KK355 pKa = 10.15EE356 pKa = 4.13KK357 pKa = 11.04ASSAGSEE364 pKa = 4.29TWWQSTKK371 pKa = 10.43FDD373 pKa = 3.84FWWYY377 pKa = 10.22NATSRR382 pKa = 11.84SWYY385 pKa = 9.98KK386 pKa = 8.56EE387 pKa = 3.44QKK389 pKa = 9.5EE390 pKa = 3.99RR391 pKa = 11.84LVGKK395 pKa = 9.85KK396 pKa = 9.74AQASSWFGRR405 pKa = 11.84LKK407 pKa = 10.72LRR409 pKa = 11.84AEE411 pKa = 4.16SLRR414 pKa = 11.84LYY416 pKa = 9.98IVSFWHH422 pKa = 6.32WGSSVVKK429 pKa = 9.84TDD431 pKa = 2.81
Molecular weight: 48.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1954 |
431 |
1523 |
977.0 |
110.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.165 ± 1.896 | 0.768 ± 0.346 |
5.22 ± 0.471 | 6.192 ± 1.916 |
4.401 ± 0.624 | 5.681 ± 0.574 |
2.098 ± 0.737 | 4.708 ± 0.763 |
6.551 ± 1.963 | 11.617 ± 0.635 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.149 ± 0.028 | 3.582 ± 0.163 |
3.941 ± 0.626 | 3.224 ± 0.429 |
5.885 ± 0.066 | 8.751 ± 0.448 |
5.067 ± 0.297 | 7.216 ± 1.371 |
2.661 ± 0.154 | 3.122 ± 0.257 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
