
Dinoponera quadriceps (South American ant)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Hymenoptera; Apocrita; Aculeata; Formicoidea; Formicidae; Ponerinae;
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
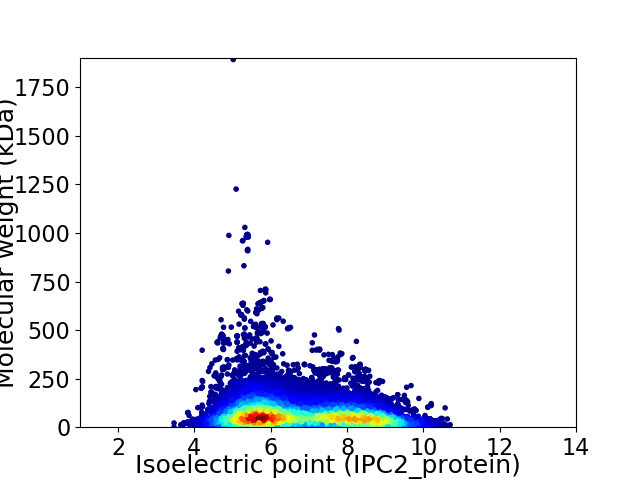
Virtual 2D-PAGE plot for 17673 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P3YH17|A0A6P3YH17_DINQU homer protein homolog 2-like OS=Dinoponera quadriceps OX=609295 GN=LOC106752136 PE=4 SV=1
MM1 pKa = 7.11VVLSNFLTPQEE12 pKa = 4.6GIRR15 pKa = 11.84HH16 pKa = 5.62EE17 pKa = 4.32EE18 pKa = 3.96QNTTVYY24 pKa = 10.76INDD27 pKa = 3.7RR28 pKa = 11.84EE29 pKa = 4.35VGKK32 pKa = 8.78GTFYY36 pKa = 9.91ITEE39 pKa = 4.36SVLSWVNSDD48 pKa = 3.51TQQGFSLEE56 pKa = 4.16YY57 pKa = 10.02PHH59 pKa = 7.27ISLHH63 pKa = 6.32AISRR67 pKa = 11.84DD68 pKa = 3.7EE69 pKa = 3.97QVHH72 pKa = 5.92PRR74 pKa = 11.84QCLYY78 pKa = 11.52VMVDD82 pKa = 3.56GKK84 pKa = 11.37VDD86 pKa = 4.07LPDD89 pKa = 3.53MPLPLSPDD97 pKa = 3.18NGSEE101 pKa = 4.29NEE103 pKa = 4.72DD104 pKa = 4.13DD105 pKa = 6.19DD106 pKa = 6.56GDD108 pKa = 4.1TPITEE113 pKa = 4.07MRR115 pKa = 11.84FAPDD119 pKa = 3.05NTNNLEE125 pKa = 4.9LMFQAMNACQALHH138 pKa = 7.51PDD140 pKa = 3.96PQDD143 pKa = 3.33SFSDD147 pKa = 3.31EE148 pKa = 5.23DD149 pKa = 3.62IYY151 pKa = 11.56EE152 pKa = 4.56DD153 pKa = 3.43AAEE156 pKa = 4.96DD157 pKa = 3.49YY158 pKa = 11.34GEE160 pKa = 4.04YY161 pKa = 10.26SEE163 pKa = 5.55EE164 pKa = 4.17VGAGDD169 pKa = 4.62APYY172 pKa = 10.4ILPTEE177 pKa = 4.13QMGASHH183 pKa = 6.78NGSEE187 pKa = 4.58AEE189 pKa = 3.84EE190 pKa = 4.32AMDD193 pKa = 3.75VEE195 pKa = 4.86AGQFEE200 pKa = 4.57DD201 pKa = 5.56AEE203 pKa = 4.32EE204 pKa = 4.4DD205 pKa = 3.54PP206 pKa = 5.23
MM1 pKa = 7.11VVLSNFLTPQEE12 pKa = 4.6GIRR15 pKa = 11.84HH16 pKa = 5.62EE17 pKa = 4.32EE18 pKa = 3.96QNTTVYY24 pKa = 10.76INDD27 pKa = 3.7RR28 pKa = 11.84EE29 pKa = 4.35VGKK32 pKa = 8.78GTFYY36 pKa = 9.91ITEE39 pKa = 4.36SVLSWVNSDD48 pKa = 3.51TQQGFSLEE56 pKa = 4.16YY57 pKa = 10.02PHH59 pKa = 7.27ISLHH63 pKa = 6.32AISRR67 pKa = 11.84DD68 pKa = 3.7EE69 pKa = 3.97QVHH72 pKa = 5.92PRR74 pKa = 11.84QCLYY78 pKa = 11.52VMVDD82 pKa = 3.56GKK84 pKa = 11.37VDD86 pKa = 4.07LPDD89 pKa = 3.53MPLPLSPDD97 pKa = 3.18NGSEE101 pKa = 4.29NEE103 pKa = 4.72DD104 pKa = 4.13DD105 pKa = 6.19DD106 pKa = 6.56GDD108 pKa = 4.1TPITEE113 pKa = 4.07MRR115 pKa = 11.84FAPDD119 pKa = 3.05NTNNLEE125 pKa = 4.9LMFQAMNACQALHH138 pKa = 7.51PDD140 pKa = 3.96PQDD143 pKa = 3.33SFSDD147 pKa = 3.31EE148 pKa = 5.23DD149 pKa = 3.62IYY151 pKa = 11.56EE152 pKa = 4.56DD153 pKa = 3.43AAEE156 pKa = 4.96DD157 pKa = 3.49YY158 pKa = 11.34GEE160 pKa = 4.04YY161 pKa = 10.26SEE163 pKa = 5.55EE164 pKa = 4.17VGAGDD169 pKa = 4.62APYY172 pKa = 10.4ILPTEE177 pKa = 4.13QMGASHH183 pKa = 6.78NGSEE187 pKa = 4.58AEE189 pKa = 3.84EE190 pKa = 4.32AMDD193 pKa = 3.75VEE195 pKa = 4.86AGQFEE200 pKa = 4.57DD201 pKa = 5.56AEE203 pKa = 4.32EE204 pKa = 4.4DD205 pKa = 3.54PP206 pKa = 5.23
Molecular weight: 22.97 kDa
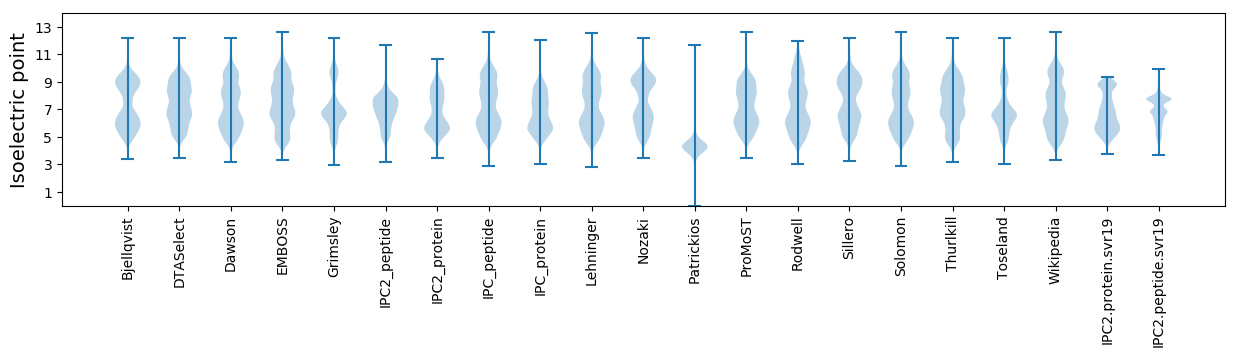
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P3XQ50|A0A6P3XQ50_DINQU zinc finger protein 346-like OS=Dinoponera quadriceps OX=609295 GN=LOC106747538 PE=4 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 9.89CGVEE7 pKa = 3.87KK8 pKa = 9.09PTDD11 pKa = 3.51VTIEE15 pKa = 4.04CSSRR19 pKa = 11.84PRR21 pKa = 11.84VNHH24 pKa = 6.49FDD26 pKa = 3.21RR27 pKa = 11.84TALVQRR33 pKa = 11.84CDD35 pKa = 3.68FRR37 pKa = 11.84SHH39 pKa = 5.71VLSTSRR45 pKa = 11.84DD46 pKa = 3.71PIRR49 pKa = 11.84NSSNRR54 pKa = 11.84QLRR57 pKa = 11.84SRR59 pKa = 11.84NGEE62 pKa = 3.76ITSSGDD68 pKa = 3.0HH69 pKa = 6.24TNPLFSVSDD78 pKa = 3.45AGQRR82 pKa = 11.84PTWSLPLSRR91 pKa = 11.84YY92 pKa = 8.92GSPFPRR98 pKa = 11.84GPSSSLDD105 pKa = 3.44PLLPFPPGEE114 pKa = 4.25SARR117 pKa = 11.84SRR119 pKa = 11.84IFIPITEE126 pKa = 4.46PDD128 pKa = 3.19SRR130 pKa = 11.84SCFRR134 pKa = 11.84CCRR137 pKa = 11.84RR138 pKa = 11.84SPPEE142 pKa = 3.74RR143 pKa = 11.84LVLCGSTALFLNHH156 pKa = 6.54SPTCAVCHH164 pKa = 6.26DD165 pKa = 4.13VGPPRR170 pKa = 11.84LLLSLSRR177 pKa = 11.84FKK179 pKa = 10.82PSTVIRR185 pKa = 11.84DD186 pKa = 3.83DD187 pKa = 4.65DD188 pKa = 4.04NGASSGQEE196 pKa = 2.84RR197 pKa = 11.84RR198 pKa = 11.84TEE200 pKa = 3.82MEE202 pKa = 3.74RR203 pKa = 11.84KK204 pKa = 9.33VKK206 pKa = 10.21IEE208 pKa = 3.97RR209 pKa = 11.84EE210 pKa = 4.01VHH212 pKa = 5.41HH213 pKa = 6.48VGKK216 pKa = 10.41IAGTKK221 pKa = 9.1ARR223 pKa = 11.84YY224 pKa = 8.92VLAVSVNKK232 pKa = 10.54LKK234 pKa = 10.89RR235 pKa = 11.84HH236 pKa = 5.48FAYY239 pKa = 10.33ASAQHH244 pKa = 6.15KK245 pKa = 9.59RR246 pKa = 11.84RR247 pKa = 11.84KK248 pKa = 7.73TRR250 pKa = 11.84ALTHH254 pKa = 6.32RR255 pKa = 11.84PARR258 pKa = 11.84SYY260 pKa = 9.6TQLNARR266 pKa = 11.84TYY268 pKa = 8.6AHH270 pKa = 6.5TRR272 pKa = 11.84NKK274 pKa = 10.05HH275 pKa = 5.15AGATIDD281 pKa = 3.54FTVCLRR287 pKa = 11.84SLSLSLSLFSFLFEE301 pKa = 4.04EE302 pKa = 4.64LHH304 pKa = 5.8AHH306 pKa = 6.48GARR309 pKa = 11.84FF310 pKa = 3.27
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 9.89CGVEE7 pKa = 3.87KK8 pKa = 9.09PTDD11 pKa = 3.51VTIEE15 pKa = 4.04CSSRR19 pKa = 11.84PRR21 pKa = 11.84VNHH24 pKa = 6.49FDD26 pKa = 3.21RR27 pKa = 11.84TALVQRR33 pKa = 11.84CDD35 pKa = 3.68FRR37 pKa = 11.84SHH39 pKa = 5.71VLSTSRR45 pKa = 11.84DD46 pKa = 3.71PIRR49 pKa = 11.84NSSNRR54 pKa = 11.84QLRR57 pKa = 11.84SRR59 pKa = 11.84NGEE62 pKa = 3.76ITSSGDD68 pKa = 3.0HH69 pKa = 6.24TNPLFSVSDD78 pKa = 3.45AGQRR82 pKa = 11.84PTWSLPLSRR91 pKa = 11.84YY92 pKa = 8.92GSPFPRR98 pKa = 11.84GPSSSLDD105 pKa = 3.44PLLPFPPGEE114 pKa = 4.25SARR117 pKa = 11.84SRR119 pKa = 11.84IFIPITEE126 pKa = 4.46PDD128 pKa = 3.19SRR130 pKa = 11.84SCFRR134 pKa = 11.84CCRR137 pKa = 11.84RR138 pKa = 11.84SPPEE142 pKa = 3.74RR143 pKa = 11.84LVLCGSTALFLNHH156 pKa = 6.54SPTCAVCHH164 pKa = 6.26DD165 pKa = 4.13VGPPRR170 pKa = 11.84LLLSLSRR177 pKa = 11.84FKK179 pKa = 10.82PSTVIRR185 pKa = 11.84DD186 pKa = 3.83DD187 pKa = 4.65DD188 pKa = 4.04NGASSGQEE196 pKa = 2.84RR197 pKa = 11.84RR198 pKa = 11.84TEE200 pKa = 3.82MEE202 pKa = 3.74RR203 pKa = 11.84KK204 pKa = 9.33VKK206 pKa = 10.21IEE208 pKa = 3.97RR209 pKa = 11.84EE210 pKa = 4.01VHH212 pKa = 5.41HH213 pKa = 6.48VGKK216 pKa = 10.41IAGTKK221 pKa = 9.1ARR223 pKa = 11.84YY224 pKa = 8.92VLAVSVNKK232 pKa = 10.54LKK234 pKa = 10.89RR235 pKa = 11.84HH236 pKa = 5.48FAYY239 pKa = 10.33ASAQHH244 pKa = 6.15KK245 pKa = 9.59RR246 pKa = 11.84RR247 pKa = 11.84KK248 pKa = 7.73TRR250 pKa = 11.84ALTHH254 pKa = 6.32RR255 pKa = 11.84PARR258 pKa = 11.84SYY260 pKa = 9.6TQLNARR266 pKa = 11.84TYY268 pKa = 8.6AHH270 pKa = 6.5TRR272 pKa = 11.84NKK274 pKa = 10.05HH275 pKa = 5.15AGATIDD281 pKa = 3.54FTVCLRR287 pKa = 11.84SLSLSLSLFSFLFEE301 pKa = 4.04EE302 pKa = 4.64LHH304 pKa = 5.8AHH306 pKa = 6.48GARR309 pKa = 11.84FF310 pKa = 3.27
Molecular weight: 34.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12470770 |
9 |
16744 |
705.6 |
79.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.824 ± 0.025 | 1.863 ± 0.019 |
5.625 ± 0.016 | 6.974 ± 0.032 |
3.249 ± 0.015 | 5.659 ± 0.021 |
2.519 ± 0.01 | 5.133 ± 0.015 |
6.038 ± 0.028 | 8.88 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.29 ± 0.009 | 4.646 ± 0.015 |
5.324 ± 0.024 | 4.492 ± 0.02 |
6.018 ± 0.02 | 8.358 ± 0.023 |
6.0 ± 0.015 | 6.172 ± 0.014 |
1.025 ± 0.007 | 2.911 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
