
Sanxia picorna-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
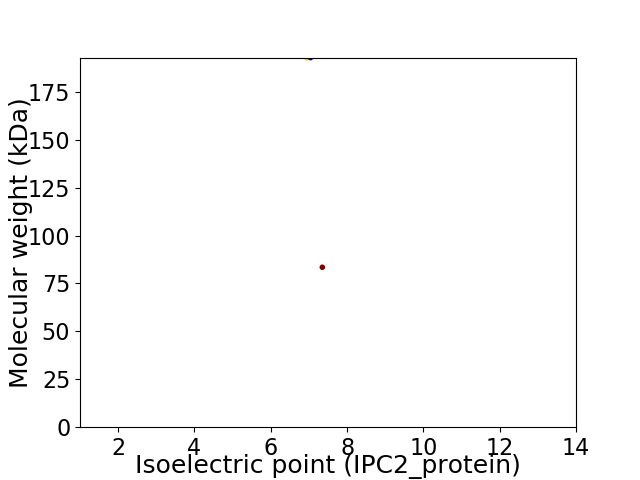
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJH8|A0A1L3KJH8_9VIRU Uncharacterized protein OS=Sanxia picorna-like virus 5 OX=1923374 PE=4 SV=1
MM1 pKa = 7.84DD2 pKa = 6.13KK3 pKa = 10.39IVNLAYY9 pKa = 7.33TTLLSFSSYY18 pKa = 10.73AVRR21 pKa = 11.84CISLAGEE28 pKa = 3.78IIEE31 pKa = 4.41RR32 pKa = 11.84IFYY35 pKa = 9.87ILYY38 pKa = 10.02FAFCRR43 pKa = 11.84MGEE46 pKa = 4.08NIDD49 pKa = 3.53FYY51 pKa = 11.78GRR53 pKa = 11.84IFVRR57 pKa = 11.84FVFNLRR63 pKa = 11.84YY64 pKa = 9.48EE65 pKa = 4.27SRR67 pKa = 11.84CFYY70 pKa = 10.85VAGGHH75 pKa = 6.13IFSSQSGYY83 pKa = 10.99AHH85 pKa = 6.64SKK87 pKa = 9.15YY88 pKa = 9.89KK89 pKa = 10.44RR90 pKa = 11.84KK91 pKa = 10.15NEE93 pKa = 3.98RR94 pKa = 11.84EE95 pKa = 3.62VSKK98 pKa = 10.52ILKK101 pKa = 9.98EE102 pKa = 3.79IGKK105 pKa = 8.89LQEE108 pKa = 4.42KK109 pKa = 10.18KK110 pKa = 10.3RR111 pKa = 11.84SCKK114 pKa = 10.65NGDD117 pKa = 3.01AKK119 pKa = 10.64KK120 pKa = 10.38YY121 pKa = 8.21YY122 pKa = 10.81EE123 pKa = 4.61PMQFLNKK130 pKa = 9.65EE131 pKa = 3.58KK132 pKa = 10.35RR133 pKa = 11.84RR134 pKa = 11.84YY135 pKa = 8.89EE136 pKa = 4.0SHH138 pKa = 6.38SGSNVRR144 pKa = 11.84PGKK147 pKa = 10.71VPLPSDD153 pKa = 3.34DD154 pKa = 3.39EE155 pKa = 4.18FRR157 pKa = 11.84IFRR160 pKa = 11.84IGLSVGFISSLWCAFFSSYY179 pKa = 10.45IFPGMMSLHH188 pKa = 5.6RR189 pKa = 11.84RR190 pKa = 11.84YY191 pKa = 9.5WDD193 pKa = 5.14DD194 pKa = 2.9ISKK197 pKa = 10.99SNLVRR202 pKa = 11.84RR203 pKa = 11.84IEE205 pKa = 3.89RR206 pKa = 11.84DD207 pKa = 2.81IPRR210 pKa = 11.84NLDD213 pKa = 3.17FLDD216 pKa = 3.63RR217 pKa = 11.84ARR219 pKa = 11.84IDD221 pKa = 3.66FSFLRR226 pKa = 11.84LATKK230 pKa = 10.26FDD232 pKa = 4.11ANNIEE237 pKa = 4.13NWTALIVGLATSTSTVNAGALLLTHH262 pKa = 7.17FKK264 pKa = 10.02TYY266 pKa = 10.19YY267 pKa = 8.61NKK269 pKa = 10.41SVAVALADD277 pKa = 3.7KK278 pKa = 10.52FSSLFMRR285 pKa = 11.84TYY287 pKa = 10.38EE288 pKa = 4.0PHH290 pKa = 6.7SLQDD294 pKa = 3.39MAGLWRR300 pKa = 11.84MLSNDD305 pKa = 3.35FNSLQKK311 pKa = 10.93SPFFDD316 pKa = 2.91KK317 pKa = 11.02VLNVVSLVVCSGLCGSFDD335 pKa = 3.46IDD337 pKa = 3.83FKK339 pKa = 11.56VSGFNVFSEE348 pKa = 4.34NLSKK352 pKa = 10.89RR353 pKa = 11.84LSSVSLTDD361 pKa = 3.36MPGMILEE368 pKa = 4.37TVAYY372 pKa = 9.24FLEE375 pKa = 4.46TGYY378 pKa = 9.04MCYY381 pKa = 7.93TQKK384 pKa = 10.8SLKK387 pKa = 10.0PILFTNPEE395 pKa = 3.93AYY397 pKa = 10.18AFEE400 pKa = 4.16QKK402 pKa = 10.47YY403 pKa = 10.52LEE405 pKa = 4.23FFRR408 pKa = 11.84VIPLIGDD415 pKa = 4.11GDD417 pKa = 3.86WEE419 pKa = 4.15AAGITISDD427 pKa = 3.37FHH429 pKa = 8.29ILYY432 pKa = 10.53DD433 pKa = 3.59EE434 pKa = 5.01LYY436 pKa = 9.87SYY438 pKa = 10.89LHH440 pKa = 6.43SLHH443 pKa = 6.84SSLNKK448 pKa = 10.16GFEE451 pKa = 4.32KK452 pKa = 10.57KK453 pKa = 10.72VIWDD457 pKa = 3.62RR458 pKa = 11.84LVNMVKK464 pKa = 9.61TKK466 pKa = 11.03NDD468 pKa = 3.52LDD470 pKa = 3.89RR471 pKa = 11.84KK472 pKa = 10.01LNSGLLRR479 pKa = 11.84RR480 pKa = 11.84APFVLAFCGPSSVGKK495 pKa = 7.0TTVANIINVVAVKK508 pKa = 10.54ASGGTGDD515 pKa = 4.01LTKK518 pKa = 10.72KK519 pKa = 8.87ITWNEE524 pKa = 3.45NDD526 pKa = 5.15DD527 pKa = 3.74YY528 pKa = 11.37FSNYY532 pKa = 9.57KK533 pKa = 10.01VDD535 pKa = 3.51TEE537 pKa = 4.57TIVMDD542 pKa = 4.67DD543 pKa = 3.83LCNTKK548 pKa = 9.96PLFIQSSPLAWLIKK562 pKa = 10.18FNNNNPEE569 pKa = 4.1YY570 pKa = 10.88AVMAEE575 pKa = 4.42LEE577 pKa = 4.7SKK579 pKa = 10.73GKK581 pKa = 10.64LPIRR585 pKa = 11.84PLTLVITTNVPDD597 pKa = 5.27LLAQTYY603 pKa = 10.21SNEE606 pKa = 3.89PVSILRR612 pKa = 11.84RR613 pKa = 11.84LDD615 pKa = 3.05MRR617 pKa = 11.84VSVTVKK623 pKa = 10.37EE624 pKa = 4.44QFSLMNGGSGNLMLDD639 pKa = 3.57PEE641 pKa = 4.39RR642 pKa = 11.84ARR644 pKa = 11.84VYY646 pKa = 10.92VDD648 pKa = 3.25SLKK651 pKa = 10.84GDD653 pKa = 3.52EE654 pKa = 5.6KK655 pKa = 10.76IFPDD659 pKa = 3.15MWNFTVEE666 pKa = 3.84KK667 pKa = 10.15AVAVKK672 pKa = 10.71NPTGGTDD679 pKa = 2.7RR680 pKa = 11.84AEE682 pKa = 3.91FRR684 pKa = 11.84KK685 pKa = 9.99VVWQNMILEE694 pKa = 4.99DD695 pKa = 3.8ISLKK699 pKa = 10.82VLADD703 pKa = 3.56YY704 pKa = 8.74TADD707 pKa = 3.49AARR710 pKa = 11.84NHH712 pKa = 6.59ANNQKK717 pKa = 10.58SLVSKK722 pKa = 8.21QTQLHH727 pKa = 5.7DD728 pKa = 4.54TINLCCVCNKK738 pKa = 10.17LQVSCQCTYY747 pKa = 10.97CDD749 pKa = 3.42QAGEE753 pKa = 4.12EE754 pKa = 4.33SSDD757 pKa = 3.82AEE759 pKa = 4.21LDD761 pKa = 3.33ASGVIDD767 pKa = 4.08ITDD770 pKa = 3.08QDD772 pKa = 4.71FIGEE776 pKa = 4.11SSFWDD781 pKa = 5.16FIPFPYY787 pKa = 10.31RR788 pKa = 11.84EE789 pKa = 4.73DD790 pKa = 2.87IAMFVSYY797 pKa = 9.71FASWRR802 pKa = 11.84GLTDD806 pKa = 2.87YY807 pKa = 11.21VRR809 pKa = 11.84LHH811 pKa = 5.72WRR813 pKa = 11.84QIFISFSLWAILSILRR829 pKa = 11.84PLNLLTCMVWCMFIFHH845 pKa = 7.31SILIYY850 pKa = 10.31RR851 pKa = 11.84FYY853 pKa = 11.03LIYY856 pKa = 9.27WQSRR860 pKa = 11.84TRR862 pKa = 11.84WFQLQSSAHH871 pKa = 6.73RR872 pKa = 11.84IASSPLFTCIVGAGTTYY889 pKa = 11.59LMTRR893 pKa = 11.84ALGKK897 pKa = 9.96ILYY900 pKa = 9.79GFIKK904 pKa = 10.21FYY906 pKa = 10.98RR907 pKa = 11.84SFNGAIMTGQSALNPDD923 pKa = 3.68GDD925 pKa = 4.17DD926 pKa = 3.44EE927 pKa = 4.7YY928 pKa = 10.89MKK930 pKa = 10.55RR931 pKa = 11.84RR932 pKa = 11.84RR933 pKa = 11.84EE934 pKa = 3.85PNLWYY939 pKa = 9.47PKK941 pKa = 10.52LPVVKK946 pKa = 10.03LAKK949 pKa = 9.72PSSRR953 pKa = 11.84SKK955 pKa = 6.78TTNWRR960 pKa = 11.84DD961 pKa = 3.46LNGKK965 pKa = 8.06IAKK968 pKa = 9.28NCVFIKK974 pKa = 10.57SGTKK978 pKa = 7.82VTGGFYY984 pKa = 10.89VRR986 pKa = 11.84THH988 pKa = 6.5LLAVPGHH995 pKa = 6.54FISPDD1000 pKa = 3.25MEE1002 pKa = 4.51LEE1004 pKa = 4.1IIPRR1008 pKa = 11.84VKK1010 pKa = 10.92YY1011 pKa = 10.64LDD1013 pKa = 3.73NKK1015 pKa = 8.99HH1016 pKa = 6.21SYY1018 pKa = 9.7RR1019 pKa = 11.84VRR1021 pKa = 11.84TNGDD1025 pKa = 3.17LVYY1028 pKa = 10.35KK1029 pKa = 10.47VPNSDD1034 pKa = 2.79IALVYY1039 pKa = 10.81APGGCDD1045 pKa = 2.66KK1046 pKa = 11.29CDD1048 pKa = 3.46FTDD1051 pKa = 3.91YY1052 pKa = 10.71FPSSHH1057 pKa = 7.22LEE1059 pKa = 4.15GEE1061 pKa = 4.81CVGSFTYY1068 pKa = 10.37RR1069 pKa = 11.84GADD1072 pKa = 3.39GEE1074 pKa = 4.41LLLDD1078 pKa = 3.99RR1079 pKa = 11.84VRR1081 pKa = 11.84MKK1083 pKa = 10.57FGKK1086 pKa = 8.66VTTDD1090 pKa = 2.66VATFSGAEE1098 pKa = 3.79YY1099 pKa = 10.59EE1100 pKa = 4.42FNNFNTFKK1108 pKa = 9.69GLCMGALVGEE1118 pKa = 4.51MKK1120 pKa = 10.12PPCIAGFHH1128 pKa = 6.47LGGVTDD1134 pKa = 3.82TQYY1137 pKa = 11.54GASGTLLKK1145 pKa = 10.13TDD1147 pKa = 3.13IRR1149 pKa = 11.84NALAHH1154 pKa = 6.52FDD1156 pKa = 3.57VPGIIFPGGSSEE1168 pKa = 4.23IPTVMYY1174 pKa = 9.91EE1175 pKa = 3.9SHH1177 pKa = 7.03LGDD1180 pKa = 4.99GPLTLDD1186 pKa = 3.59KK1187 pKa = 10.35PISPRR1192 pKa = 11.84CSSNFLPEE1200 pKa = 3.68GAIIDD1205 pKa = 3.87VLASCKK1211 pKa = 10.48GAVTNKK1217 pKa = 10.04SEE1219 pKa = 4.48VIVSHH1224 pKa = 6.61ISGKK1228 pKa = 8.78VFEE1231 pKa = 4.37FTGVEE1236 pKa = 4.17RR1237 pKa = 11.84THH1239 pKa = 6.55GVAPMGPPLVRR1250 pKa = 11.84SWHH1253 pKa = 5.26NWSLGMQGFSDD1264 pKa = 3.98PAVGPFLDD1272 pKa = 3.78SVIRR1276 pKa = 11.84ASVDD1280 pKa = 3.29YY1281 pKa = 10.44IQPLLPKK1288 pKa = 10.01FINILPLDD1296 pKa = 4.57LPTIINGLDD1305 pKa = 3.16GDD1307 pKa = 4.09KK1308 pKa = 10.66FLNRR1312 pKa = 11.84MPQNTSVGFPLSGKK1326 pKa = 9.58LSKK1329 pKa = 10.77FSTIVPAQEE1338 pKa = 3.73GHH1340 pKa = 5.64SVNFEE1345 pKa = 4.17LSPEE1349 pKa = 3.72IMEE1352 pKa = 4.13VYY1354 pKa = 10.28EE1355 pKa = 4.17IYY1357 pKa = 9.12KK1358 pKa = 8.25TRR1360 pKa = 11.84YY1361 pKa = 8.25RR1362 pKa = 11.84AGEE1365 pKa = 3.56RR1366 pKa = 11.84CYY1368 pKa = 10.55PIFRR1372 pKa = 11.84ASLKK1376 pKa = 10.63DD1377 pKa = 3.55EE1378 pKa = 4.0PVKK1381 pKa = 10.5IGKK1384 pKa = 9.68LKK1386 pKa = 10.68VRR1388 pKa = 11.84VFQAAPVALKK1398 pKa = 9.85MLLRR1402 pKa = 11.84EE1403 pKa = 4.05YY1404 pKa = 10.28FLPIASHH1411 pKa = 6.93LSMFPLISEE1420 pKa = 4.49CAVGVNAFSNEE1431 pKa = 3.68WEE1433 pKa = 4.48EE1434 pKa = 4.02MHH1436 pKa = 5.82QHH1438 pKa = 5.27IVKK1441 pKa = 10.52FGDD1444 pKa = 3.33SRR1446 pKa = 11.84IVAGDD1451 pKa = 3.15YY1452 pKa = 10.44SAYY1455 pKa = 9.29DD1456 pKa = 3.22QRR1458 pKa = 11.84MPPSLTGAAFSILIEE1473 pKa = 4.18LAEE1476 pKa = 4.23KK1477 pKa = 10.55AGYY1480 pKa = 10.14SPDD1483 pKa = 3.91DD1484 pKa = 3.74LTIMRR1489 pKa = 11.84AMVADD1494 pKa = 4.19VIYY1497 pKa = 10.27PLVAYY1502 pKa = 10.41NGTLVQFYY1510 pKa = 10.27GSNPSGHH1517 pKa = 6.43NLTVYY1522 pKa = 10.38INSIVNSLISRR1533 pKa = 11.84CAFFSMYY1540 pKa = 9.95PEE1542 pKa = 4.44HH1543 pKa = 7.44RR1544 pKa = 11.84DD1545 pKa = 3.23FRR1547 pKa = 11.84LAVSMITYY1555 pKa = 10.45GDD1557 pKa = 3.99DD1558 pKa = 4.71DD1559 pKa = 3.94IGSVSEE1565 pKa = 4.42AYY1567 pKa = 10.48GDD1569 pKa = 3.99FNCVTKK1575 pKa = 10.76SNYY1578 pKa = 9.51INYY1581 pKa = 9.42IGMKK1585 pKa = 7.79YY1586 pKa = 8.61TPPDD1590 pKa = 3.15KK1591 pKa = 10.98SGEE1594 pKa = 4.15HH1595 pKa = 4.93VPYY1598 pKa = 10.16MEE1600 pKa = 4.29ISGVDD1605 pKa = 3.69FLKK1608 pKa = 10.69RR1609 pKa = 11.84KK1610 pKa = 9.84SVFNTHH1616 pKa = 6.56LDD1618 pKa = 3.46RR1619 pKa = 11.84HH1620 pKa = 6.27IGALDD1625 pKa = 3.2KK1626 pKa = 11.4ASIYY1630 pKa = 10.46KK1631 pKa = 9.91SLHH1634 pKa = 4.04VRR1636 pKa = 11.84MRR1638 pKa = 11.84SSDD1641 pKa = 3.2ISDD1644 pKa = 3.69EE1645 pKa = 4.06AWAGAVVDD1653 pKa = 4.0GALRR1657 pKa = 11.84EE1658 pKa = 4.28FFAHH1662 pKa = 7.2GEE1664 pKa = 3.55QDD1666 pKa = 3.6YY1667 pKa = 11.19EE1668 pKa = 4.15IFRR1671 pKa = 11.84SQMVRR1676 pKa = 11.84VAEE1679 pKa = 4.37DD1680 pKa = 3.31CDD1682 pKa = 4.12FAVHH1686 pKa = 6.76SSNLNFSYY1694 pKa = 11.01KK1695 pKa = 10.63EE1696 pKa = 3.83MVEE1699 pKa = 4.05KK1700 pKa = 11.05VEE1702 pKa = 4.04PQINN1706 pKa = 3.29
MM1 pKa = 7.84DD2 pKa = 6.13KK3 pKa = 10.39IVNLAYY9 pKa = 7.33TTLLSFSSYY18 pKa = 10.73AVRR21 pKa = 11.84CISLAGEE28 pKa = 3.78IIEE31 pKa = 4.41RR32 pKa = 11.84IFYY35 pKa = 9.87ILYY38 pKa = 10.02FAFCRR43 pKa = 11.84MGEE46 pKa = 4.08NIDD49 pKa = 3.53FYY51 pKa = 11.78GRR53 pKa = 11.84IFVRR57 pKa = 11.84FVFNLRR63 pKa = 11.84YY64 pKa = 9.48EE65 pKa = 4.27SRR67 pKa = 11.84CFYY70 pKa = 10.85VAGGHH75 pKa = 6.13IFSSQSGYY83 pKa = 10.99AHH85 pKa = 6.64SKK87 pKa = 9.15YY88 pKa = 9.89KK89 pKa = 10.44RR90 pKa = 11.84KK91 pKa = 10.15NEE93 pKa = 3.98RR94 pKa = 11.84EE95 pKa = 3.62VSKK98 pKa = 10.52ILKK101 pKa = 9.98EE102 pKa = 3.79IGKK105 pKa = 8.89LQEE108 pKa = 4.42KK109 pKa = 10.18KK110 pKa = 10.3RR111 pKa = 11.84SCKK114 pKa = 10.65NGDD117 pKa = 3.01AKK119 pKa = 10.64KK120 pKa = 10.38YY121 pKa = 8.21YY122 pKa = 10.81EE123 pKa = 4.61PMQFLNKK130 pKa = 9.65EE131 pKa = 3.58KK132 pKa = 10.35RR133 pKa = 11.84RR134 pKa = 11.84YY135 pKa = 8.89EE136 pKa = 4.0SHH138 pKa = 6.38SGSNVRR144 pKa = 11.84PGKK147 pKa = 10.71VPLPSDD153 pKa = 3.34DD154 pKa = 3.39EE155 pKa = 4.18FRR157 pKa = 11.84IFRR160 pKa = 11.84IGLSVGFISSLWCAFFSSYY179 pKa = 10.45IFPGMMSLHH188 pKa = 5.6RR189 pKa = 11.84RR190 pKa = 11.84YY191 pKa = 9.5WDD193 pKa = 5.14DD194 pKa = 2.9ISKK197 pKa = 10.99SNLVRR202 pKa = 11.84RR203 pKa = 11.84IEE205 pKa = 3.89RR206 pKa = 11.84DD207 pKa = 2.81IPRR210 pKa = 11.84NLDD213 pKa = 3.17FLDD216 pKa = 3.63RR217 pKa = 11.84ARR219 pKa = 11.84IDD221 pKa = 3.66FSFLRR226 pKa = 11.84LATKK230 pKa = 10.26FDD232 pKa = 4.11ANNIEE237 pKa = 4.13NWTALIVGLATSTSTVNAGALLLTHH262 pKa = 7.17FKK264 pKa = 10.02TYY266 pKa = 10.19YY267 pKa = 8.61NKK269 pKa = 10.41SVAVALADD277 pKa = 3.7KK278 pKa = 10.52FSSLFMRR285 pKa = 11.84TYY287 pKa = 10.38EE288 pKa = 4.0PHH290 pKa = 6.7SLQDD294 pKa = 3.39MAGLWRR300 pKa = 11.84MLSNDD305 pKa = 3.35FNSLQKK311 pKa = 10.93SPFFDD316 pKa = 2.91KK317 pKa = 11.02VLNVVSLVVCSGLCGSFDD335 pKa = 3.46IDD337 pKa = 3.83FKK339 pKa = 11.56VSGFNVFSEE348 pKa = 4.34NLSKK352 pKa = 10.89RR353 pKa = 11.84LSSVSLTDD361 pKa = 3.36MPGMILEE368 pKa = 4.37TVAYY372 pKa = 9.24FLEE375 pKa = 4.46TGYY378 pKa = 9.04MCYY381 pKa = 7.93TQKK384 pKa = 10.8SLKK387 pKa = 10.0PILFTNPEE395 pKa = 3.93AYY397 pKa = 10.18AFEE400 pKa = 4.16QKK402 pKa = 10.47YY403 pKa = 10.52LEE405 pKa = 4.23FFRR408 pKa = 11.84VIPLIGDD415 pKa = 4.11GDD417 pKa = 3.86WEE419 pKa = 4.15AAGITISDD427 pKa = 3.37FHH429 pKa = 8.29ILYY432 pKa = 10.53DD433 pKa = 3.59EE434 pKa = 5.01LYY436 pKa = 9.87SYY438 pKa = 10.89LHH440 pKa = 6.43SLHH443 pKa = 6.84SSLNKK448 pKa = 10.16GFEE451 pKa = 4.32KK452 pKa = 10.57KK453 pKa = 10.72VIWDD457 pKa = 3.62RR458 pKa = 11.84LVNMVKK464 pKa = 9.61TKK466 pKa = 11.03NDD468 pKa = 3.52LDD470 pKa = 3.89RR471 pKa = 11.84KK472 pKa = 10.01LNSGLLRR479 pKa = 11.84RR480 pKa = 11.84APFVLAFCGPSSVGKK495 pKa = 7.0TTVANIINVVAVKK508 pKa = 10.54ASGGTGDD515 pKa = 4.01LTKK518 pKa = 10.72KK519 pKa = 8.87ITWNEE524 pKa = 3.45NDD526 pKa = 5.15DD527 pKa = 3.74YY528 pKa = 11.37FSNYY532 pKa = 9.57KK533 pKa = 10.01VDD535 pKa = 3.51TEE537 pKa = 4.57TIVMDD542 pKa = 4.67DD543 pKa = 3.83LCNTKK548 pKa = 9.96PLFIQSSPLAWLIKK562 pKa = 10.18FNNNNPEE569 pKa = 4.1YY570 pKa = 10.88AVMAEE575 pKa = 4.42LEE577 pKa = 4.7SKK579 pKa = 10.73GKK581 pKa = 10.64LPIRR585 pKa = 11.84PLTLVITTNVPDD597 pKa = 5.27LLAQTYY603 pKa = 10.21SNEE606 pKa = 3.89PVSILRR612 pKa = 11.84RR613 pKa = 11.84LDD615 pKa = 3.05MRR617 pKa = 11.84VSVTVKK623 pKa = 10.37EE624 pKa = 4.44QFSLMNGGSGNLMLDD639 pKa = 3.57PEE641 pKa = 4.39RR642 pKa = 11.84ARR644 pKa = 11.84VYY646 pKa = 10.92VDD648 pKa = 3.25SLKK651 pKa = 10.84GDD653 pKa = 3.52EE654 pKa = 5.6KK655 pKa = 10.76IFPDD659 pKa = 3.15MWNFTVEE666 pKa = 3.84KK667 pKa = 10.15AVAVKK672 pKa = 10.71NPTGGTDD679 pKa = 2.7RR680 pKa = 11.84AEE682 pKa = 3.91FRR684 pKa = 11.84KK685 pKa = 9.99VVWQNMILEE694 pKa = 4.99DD695 pKa = 3.8ISLKK699 pKa = 10.82VLADD703 pKa = 3.56YY704 pKa = 8.74TADD707 pKa = 3.49AARR710 pKa = 11.84NHH712 pKa = 6.59ANNQKK717 pKa = 10.58SLVSKK722 pKa = 8.21QTQLHH727 pKa = 5.7DD728 pKa = 4.54TINLCCVCNKK738 pKa = 10.17LQVSCQCTYY747 pKa = 10.97CDD749 pKa = 3.42QAGEE753 pKa = 4.12EE754 pKa = 4.33SSDD757 pKa = 3.82AEE759 pKa = 4.21LDD761 pKa = 3.33ASGVIDD767 pKa = 4.08ITDD770 pKa = 3.08QDD772 pKa = 4.71FIGEE776 pKa = 4.11SSFWDD781 pKa = 5.16FIPFPYY787 pKa = 10.31RR788 pKa = 11.84EE789 pKa = 4.73DD790 pKa = 2.87IAMFVSYY797 pKa = 9.71FASWRR802 pKa = 11.84GLTDD806 pKa = 2.87YY807 pKa = 11.21VRR809 pKa = 11.84LHH811 pKa = 5.72WRR813 pKa = 11.84QIFISFSLWAILSILRR829 pKa = 11.84PLNLLTCMVWCMFIFHH845 pKa = 7.31SILIYY850 pKa = 10.31RR851 pKa = 11.84FYY853 pKa = 11.03LIYY856 pKa = 9.27WQSRR860 pKa = 11.84TRR862 pKa = 11.84WFQLQSSAHH871 pKa = 6.73RR872 pKa = 11.84IASSPLFTCIVGAGTTYY889 pKa = 11.59LMTRR893 pKa = 11.84ALGKK897 pKa = 9.96ILYY900 pKa = 9.79GFIKK904 pKa = 10.21FYY906 pKa = 10.98RR907 pKa = 11.84SFNGAIMTGQSALNPDD923 pKa = 3.68GDD925 pKa = 4.17DD926 pKa = 3.44EE927 pKa = 4.7YY928 pKa = 10.89MKK930 pKa = 10.55RR931 pKa = 11.84RR932 pKa = 11.84RR933 pKa = 11.84EE934 pKa = 3.85PNLWYY939 pKa = 9.47PKK941 pKa = 10.52LPVVKK946 pKa = 10.03LAKK949 pKa = 9.72PSSRR953 pKa = 11.84SKK955 pKa = 6.78TTNWRR960 pKa = 11.84DD961 pKa = 3.46LNGKK965 pKa = 8.06IAKK968 pKa = 9.28NCVFIKK974 pKa = 10.57SGTKK978 pKa = 7.82VTGGFYY984 pKa = 10.89VRR986 pKa = 11.84THH988 pKa = 6.5LLAVPGHH995 pKa = 6.54FISPDD1000 pKa = 3.25MEE1002 pKa = 4.51LEE1004 pKa = 4.1IIPRR1008 pKa = 11.84VKK1010 pKa = 10.92YY1011 pKa = 10.64LDD1013 pKa = 3.73NKK1015 pKa = 8.99HH1016 pKa = 6.21SYY1018 pKa = 9.7RR1019 pKa = 11.84VRR1021 pKa = 11.84TNGDD1025 pKa = 3.17LVYY1028 pKa = 10.35KK1029 pKa = 10.47VPNSDD1034 pKa = 2.79IALVYY1039 pKa = 10.81APGGCDD1045 pKa = 2.66KK1046 pKa = 11.29CDD1048 pKa = 3.46FTDD1051 pKa = 3.91YY1052 pKa = 10.71FPSSHH1057 pKa = 7.22LEE1059 pKa = 4.15GEE1061 pKa = 4.81CVGSFTYY1068 pKa = 10.37RR1069 pKa = 11.84GADD1072 pKa = 3.39GEE1074 pKa = 4.41LLLDD1078 pKa = 3.99RR1079 pKa = 11.84VRR1081 pKa = 11.84MKK1083 pKa = 10.57FGKK1086 pKa = 8.66VTTDD1090 pKa = 2.66VATFSGAEE1098 pKa = 3.79YY1099 pKa = 10.59EE1100 pKa = 4.42FNNFNTFKK1108 pKa = 9.69GLCMGALVGEE1118 pKa = 4.51MKK1120 pKa = 10.12PPCIAGFHH1128 pKa = 6.47LGGVTDD1134 pKa = 3.82TQYY1137 pKa = 11.54GASGTLLKK1145 pKa = 10.13TDD1147 pKa = 3.13IRR1149 pKa = 11.84NALAHH1154 pKa = 6.52FDD1156 pKa = 3.57VPGIIFPGGSSEE1168 pKa = 4.23IPTVMYY1174 pKa = 9.91EE1175 pKa = 3.9SHH1177 pKa = 7.03LGDD1180 pKa = 4.99GPLTLDD1186 pKa = 3.59KK1187 pKa = 10.35PISPRR1192 pKa = 11.84CSSNFLPEE1200 pKa = 3.68GAIIDD1205 pKa = 3.87VLASCKK1211 pKa = 10.48GAVTNKK1217 pKa = 10.04SEE1219 pKa = 4.48VIVSHH1224 pKa = 6.61ISGKK1228 pKa = 8.78VFEE1231 pKa = 4.37FTGVEE1236 pKa = 4.17RR1237 pKa = 11.84THH1239 pKa = 6.55GVAPMGPPLVRR1250 pKa = 11.84SWHH1253 pKa = 5.26NWSLGMQGFSDD1264 pKa = 3.98PAVGPFLDD1272 pKa = 3.78SVIRR1276 pKa = 11.84ASVDD1280 pKa = 3.29YY1281 pKa = 10.44IQPLLPKK1288 pKa = 10.01FINILPLDD1296 pKa = 4.57LPTIINGLDD1305 pKa = 3.16GDD1307 pKa = 4.09KK1308 pKa = 10.66FLNRR1312 pKa = 11.84MPQNTSVGFPLSGKK1326 pKa = 9.58LSKK1329 pKa = 10.77FSTIVPAQEE1338 pKa = 3.73GHH1340 pKa = 5.64SVNFEE1345 pKa = 4.17LSPEE1349 pKa = 3.72IMEE1352 pKa = 4.13VYY1354 pKa = 10.28EE1355 pKa = 4.17IYY1357 pKa = 9.12KK1358 pKa = 8.25TRR1360 pKa = 11.84YY1361 pKa = 8.25RR1362 pKa = 11.84AGEE1365 pKa = 3.56RR1366 pKa = 11.84CYY1368 pKa = 10.55PIFRR1372 pKa = 11.84ASLKK1376 pKa = 10.63DD1377 pKa = 3.55EE1378 pKa = 4.0PVKK1381 pKa = 10.5IGKK1384 pKa = 9.68LKK1386 pKa = 10.68VRR1388 pKa = 11.84VFQAAPVALKK1398 pKa = 9.85MLLRR1402 pKa = 11.84EE1403 pKa = 4.05YY1404 pKa = 10.28FLPIASHH1411 pKa = 6.93LSMFPLISEE1420 pKa = 4.49CAVGVNAFSNEE1431 pKa = 3.68WEE1433 pKa = 4.48EE1434 pKa = 4.02MHH1436 pKa = 5.82QHH1438 pKa = 5.27IVKK1441 pKa = 10.52FGDD1444 pKa = 3.33SRR1446 pKa = 11.84IVAGDD1451 pKa = 3.15YY1452 pKa = 10.44SAYY1455 pKa = 9.29DD1456 pKa = 3.22QRR1458 pKa = 11.84MPPSLTGAAFSILIEE1473 pKa = 4.18LAEE1476 pKa = 4.23KK1477 pKa = 10.55AGYY1480 pKa = 10.14SPDD1483 pKa = 3.91DD1484 pKa = 3.74LTIMRR1489 pKa = 11.84AMVADD1494 pKa = 4.19VIYY1497 pKa = 10.27PLVAYY1502 pKa = 10.41NGTLVQFYY1510 pKa = 10.27GSNPSGHH1517 pKa = 6.43NLTVYY1522 pKa = 10.38INSIVNSLISRR1533 pKa = 11.84CAFFSMYY1540 pKa = 9.95PEE1542 pKa = 4.44HH1543 pKa = 7.44RR1544 pKa = 11.84DD1545 pKa = 3.23FRR1547 pKa = 11.84LAVSMITYY1555 pKa = 10.45GDD1557 pKa = 3.99DD1558 pKa = 4.71DD1559 pKa = 3.94IGSVSEE1565 pKa = 4.42AYY1567 pKa = 10.48GDD1569 pKa = 3.99FNCVTKK1575 pKa = 10.76SNYY1578 pKa = 9.51INYY1581 pKa = 9.42IGMKK1585 pKa = 7.79YY1586 pKa = 8.61TPPDD1590 pKa = 3.15KK1591 pKa = 10.98SGEE1594 pKa = 4.15HH1595 pKa = 4.93VPYY1598 pKa = 10.16MEE1600 pKa = 4.29ISGVDD1605 pKa = 3.69FLKK1608 pKa = 10.69RR1609 pKa = 11.84KK1610 pKa = 9.84SVFNTHH1616 pKa = 6.56LDD1618 pKa = 3.46RR1619 pKa = 11.84HH1620 pKa = 6.27IGALDD1625 pKa = 3.2KK1626 pKa = 11.4ASIYY1630 pKa = 10.46KK1631 pKa = 9.91SLHH1634 pKa = 4.04VRR1636 pKa = 11.84MRR1638 pKa = 11.84SSDD1641 pKa = 3.2ISDD1644 pKa = 3.69EE1645 pKa = 4.06AWAGAVVDD1653 pKa = 4.0GALRR1657 pKa = 11.84EE1658 pKa = 4.28FFAHH1662 pKa = 7.2GEE1664 pKa = 3.55QDD1666 pKa = 3.6YY1667 pKa = 11.19EE1668 pKa = 4.15IFRR1671 pKa = 11.84SQMVRR1676 pKa = 11.84VAEE1679 pKa = 4.37DD1680 pKa = 3.31CDD1682 pKa = 4.12FAVHH1686 pKa = 6.76SSNLNFSYY1694 pKa = 11.01KK1695 pKa = 10.63EE1696 pKa = 3.83MVEE1699 pKa = 4.05KK1700 pKa = 11.05VEE1702 pKa = 4.04PQINN1706 pKa = 3.29
Molecular weight: 192.7 kDa
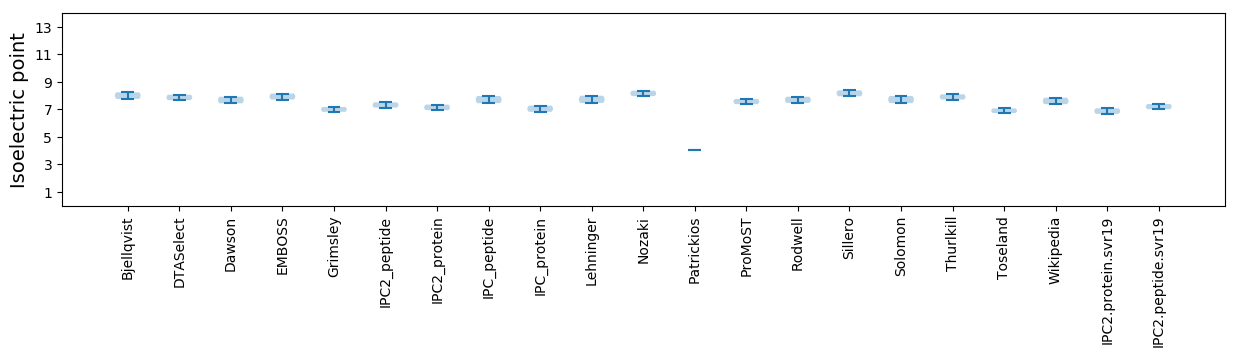
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJH8|A0A1L3KJH8_9VIRU Uncharacterized protein OS=Sanxia picorna-like virus 5 OX=1923374 PE=4 SV=1
MM1 pKa = 7.51FDD3 pKa = 2.93VGKK6 pKa = 10.32FFSRR10 pKa = 11.84PVRR13 pKa = 11.84IGSFPWTVNTAFVGQSIFPFEE34 pKa = 4.74LFWKK38 pKa = 10.07NKK40 pKa = 9.39HH41 pKa = 5.41NANRR45 pKa = 11.84LNNYY49 pKa = 10.04RR50 pKa = 11.84NMKK53 pKa = 10.27CDD55 pKa = 3.27MCVKK59 pKa = 9.91IVVNGTPFHH68 pKa = 7.07YY69 pKa = 10.64GMILASVTPDD79 pKa = 2.96QSLEE83 pKa = 3.99GFPNSDD89 pKa = 3.46LTEE92 pKa = 4.47LGCVRR97 pKa = 11.84GSQNPHH103 pKa = 6.55IFIDD107 pKa = 3.97ASTSSAGCLRR117 pKa = 11.84LPYY120 pKa = 8.48THH122 pKa = 7.72PSNAFDD128 pKa = 3.74TLGTNLQSAGTLALRR143 pKa = 11.84EE144 pKa = 4.47LVPLAHH150 pKa = 6.26VSGVVEE156 pKa = 4.8TITVSVFAWAEE167 pKa = 4.03NVVLGAPTINNISGLTAQSSDD188 pKa = 3.34EE189 pKa = 4.09YY190 pKa = 9.96GQGVISRR197 pKa = 11.84PAFALAAVAKK207 pKa = 10.56SMVKK211 pKa = 10.31FPYY214 pKa = 9.5IRR216 pKa = 11.84PYY218 pKa = 11.87AMATDD223 pKa = 4.06MTATSIGNLASLFGFVKK240 pKa = 10.41PNVVSDD246 pKa = 3.5ITLMNPRR253 pKa = 11.84ALPNLASATQHH264 pKa = 6.82DD265 pKa = 5.45PIFKK269 pKa = 9.34ATLDD273 pKa = 3.82DD274 pKa = 4.13KK275 pKa = 11.56QGVTVDD281 pKa = 3.46PRR283 pKa = 11.84VVGFDD288 pKa = 3.33GRR290 pKa = 11.84DD291 pKa = 3.12EE292 pKa = 4.1MGILDD297 pKa = 3.26IAMRR301 pKa = 11.84EE302 pKa = 4.14SYY304 pKa = 10.59LFQFPYY310 pKa = 9.49TQTNAVDD317 pKa = 3.53SRR319 pKa = 11.84LCVLTISPNQYY330 pKa = 7.91RR331 pKa = 11.84TSGTGINTQFIQTPMSWVSAPFQYY355 pKa = 9.76WRR357 pKa = 11.84GSIRR361 pKa = 11.84YY362 pKa = 8.74KK363 pKa = 10.1LTAVCSAFHH372 pKa = 7.07RR373 pKa = 11.84GRR375 pKa = 11.84LRR377 pKa = 11.84IAYY380 pKa = 7.93EE381 pKa = 3.99PAGSTAGLVPSFNVVMSEE399 pKa = 3.24IWDD402 pKa = 3.3ISEE405 pKa = 4.06KK406 pKa = 10.67KK407 pKa = 10.49EE408 pKa = 3.73IVIDD412 pKa = 4.14VGWHH416 pKa = 4.24QPNPYY421 pKa = 8.45MQVSNIGDD429 pKa = 4.61GINPFVIGVTGNIVANPFRR448 pKa = 11.84NGTISVYY455 pKa = 10.61VLNEE459 pKa = 3.67LTSPDD464 pKa = 3.56TSLTSPISILVSVSACEE481 pKa = 4.02DD482 pKa = 3.54FEE484 pKa = 4.9LFSPIDD490 pKa = 3.68TISNFTYY497 pKa = 9.61HH498 pKa = 6.05AQSGEE503 pKa = 3.93VLEE506 pKa = 4.3NSIIKK511 pKa = 9.59PYY513 pKa = 10.37KK514 pKa = 10.1SEE516 pKa = 4.38ADD518 pKa = 3.4VKK520 pKa = 9.93FGSRR524 pKa = 11.84IRR526 pKa = 11.84SNDD529 pKa = 3.21PSPKK533 pKa = 9.16IFHH536 pKa = 7.17GDD538 pKa = 3.35PVISLRR544 pKa = 11.84TLLKK548 pKa = 10.37RR549 pKa = 11.84YY550 pKa = 7.25TYY552 pKa = 10.72SCAYY556 pKa = 10.0SPTFTAVGVQRR567 pKa = 11.84FSWRR571 pKa = 11.84LVLPQMPLYY580 pKa = 9.89PGRR583 pKa = 11.84APLAVHH589 pKa = 6.55TATTAINFSSMTLLNYY605 pKa = 7.61FTPAFLARR613 pKa = 11.84RR614 pKa = 11.84GGIRR618 pKa = 11.84HH619 pKa = 6.24RR620 pKa = 11.84FIAGIGCPIEE630 pKa = 3.9HH631 pKa = 6.69AAIYY635 pKa = 7.77RR636 pKa = 11.84APVAGYY642 pKa = 8.54TNTTTLFPAITTPSVAARR660 pKa = 11.84AHH662 pKa = 5.42RR663 pKa = 11.84VEE665 pKa = 4.36HH666 pKa = 6.44RR667 pKa = 11.84GVTNGVSGMSATPDD681 pKa = 3.08IEE683 pKa = 5.4RR684 pKa = 11.84IPLDD688 pKa = 3.29IEE690 pKa = 4.44VPFYY694 pKa = 11.34SPLRR698 pKa = 11.84YY699 pKa = 9.33FRR701 pKa = 11.84GRR703 pKa = 11.84DD704 pKa = 3.01GGTNRR709 pKa = 11.84GSSSGAGVTILVKK722 pKa = 10.47FVTSSGSDD730 pKa = 3.43VILEE734 pKa = 4.32QYY736 pKa = 10.68VSAADD741 pKa = 3.81DD742 pKa = 3.94FSLHH746 pKa = 6.02GFIDD750 pKa = 3.91VPPMYY755 pKa = 11.02NHH757 pKa = 5.82TTVPAATT764 pKa = 3.93
MM1 pKa = 7.51FDD3 pKa = 2.93VGKK6 pKa = 10.32FFSRR10 pKa = 11.84PVRR13 pKa = 11.84IGSFPWTVNTAFVGQSIFPFEE34 pKa = 4.74LFWKK38 pKa = 10.07NKK40 pKa = 9.39HH41 pKa = 5.41NANRR45 pKa = 11.84LNNYY49 pKa = 10.04RR50 pKa = 11.84NMKK53 pKa = 10.27CDD55 pKa = 3.27MCVKK59 pKa = 9.91IVVNGTPFHH68 pKa = 7.07YY69 pKa = 10.64GMILASVTPDD79 pKa = 2.96QSLEE83 pKa = 3.99GFPNSDD89 pKa = 3.46LTEE92 pKa = 4.47LGCVRR97 pKa = 11.84GSQNPHH103 pKa = 6.55IFIDD107 pKa = 3.97ASTSSAGCLRR117 pKa = 11.84LPYY120 pKa = 8.48THH122 pKa = 7.72PSNAFDD128 pKa = 3.74TLGTNLQSAGTLALRR143 pKa = 11.84EE144 pKa = 4.47LVPLAHH150 pKa = 6.26VSGVVEE156 pKa = 4.8TITVSVFAWAEE167 pKa = 4.03NVVLGAPTINNISGLTAQSSDD188 pKa = 3.34EE189 pKa = 4.09YY190 pKa = 9.96GQGVISRR197 pKa = 11.84PAFALAAVAKK207 pKa = 10.56SMVKK211 pKa = 10.31FPYY214 pKa = 9.5IRR216 pKa = 11.84PYY218 pKa = 11.87AMATDD223 pKa = 4.06MTATSIGNLASLFGFVKK240 pKa = 10.41PNVVSDD246 pKa = 3.5ITLMNPRR253 pKa = 11.84ALPNLASATQHH264 pKa = 6.82DD265 pKa = 5.45PIFKK269 pKa = 9.34ATLDD273 pKa = 3.82DD274 pKa = 4.13KK275 pKa = 11.56QGVTVDD281 pKa = 3.46PRR283 pKa = 11.84VVGFDD288 pKa = 3.33GRR290 pKa = 11.84DD291 pKa = 3.12EE292 pKa = 4.1MGILDD297 pKa = 3.26IAMRR301 pKa = 11.84EE302 pKa = 4.14SYY304 pKa = 10.59LFQFPYY310 pKa = 9.49TQTNAVDD317 pKa = 3.53SRR319 pKa = 11.84LCVLTISPNQYY330 pKa = 7.91RR331 pKa = 11.84TSGTGINTQFIQTPMSWVSAPFQYY355 pKa = 9.76WRR357 pKa = 11.84GSIRR361 pKa = 11.84YY362 pKa = 8.74KK363 pKa = 10.1LTAVCSAFHH372 pKa = 7.07RR373 pKa = 11.84GRR375 pKa = 11.84LRR377 pKa = 11.84IAYY380 pKa = 7.93EE381 pKa = 3.99PAGSTAGLVPSFNVVMSEE399 pKa = 3.24IWDD402 pKa = 3.3ISEE405 pKa = 4.06KK406 pKa = 10.67KK407 pKa = 10.49EE408 pKa = 3.73IVIDD412 pKa = 4.14VGWHH416 pKa = 4.24QPNPYY421 pKa = 8.45MQVSNIGDD429 pKa = 4.61GINPFVIGVTGNIVANPFRR448 pKa = 11.84NGTISVYY455 pKa = 10.61VLNEE459 pKa = 3.67LTSPDD464 pKa = 3.56TSLTSPISILVSVSACEE481 pKa = 4.02DD482 pKa = 3.54FEE484 pKa = 4.9LFSPIDD490 pKa = 3.68TISNFTYY497 pKa = 9.61HH498 pKa = 6.05AQSGEE503 pKa = 3.93VLEE506 pKa = 4.3NSIIKK511 pKa = 9.59PYY513 pKa = 10.37KK514 pKa = 10.1SEE516 pKa = 4.38ADD518 pKa = 3.4VKK520 pKa = 9.93FGSRR524 pKa = 11.84IRR526 pKa = 11.84SNDD529 pKa = 3.21PSPKK533 pKa = 9.16IFHH536 pKa = 7.17GDD538 pKa = 3.35PVISLRR544 pKa = 11.84TLLKK548 pKa = 10.37RR549 pKa = 11.84YY550 pKa = 7.25TYY552 pKa = 10.72SCAYY556 pKa = 10.0SPTFTAVGVQRR567 pKa = 11.84FSWRR571 pKa = 11.84LVLPQMPLYY580 pKa = 9.89PGRR583 pKa = 11.84APLAVHH589 pKa = 6.55TATTAINFSSMTLLNYY605 pKa = 7.61FTPAFLARR613 pKa = 11.84RR614 pKa = 11.84GGIRR618 pKa = 11.84HH619 pKa = 6.24RR620 pKa = 11.84FIAGIGCPIEE630 pKa = 3.9HH631 pKa = 6.69AAIYY635 pKa = 7.77RR636 pKa = 11.84APVAGYY642 pKa = 8.54TNTTTLFPAITTPSVAARR660 pKa = 11.84AHH662 pKa = 5.42RR663 pKa = 11.84VEE665 pKa = 4.36HH666 pKa = 6.44RR667 pKa = 11.84GVTNGVSGMSATPDD681 pKa = 3.08IEE683 pKa = 5.4RR684 pKa = 11.84IPLDD688 pKa = 3.29IEE690 pKa = 4.44VPFYY694 pKa = 11.34SPLRR698 pKa = 11.84YY699 pKa = 9.33FRR701 pKa = 11.84GRR703 pKa = 11.84DD704 pKa = 3.01GGTNRR709 pKa = 11.84GSSSGAGVTILVKK722 pKa = 10.47FVTSSGSDD730 pKa = 3.43VILEE734 pKa = 4.32QYY736 pKa = 10.68VSAADD741 pKa = 3.81DD742 pKa = 3.94FSLHH746 pKa = 6.02GFIDD750 pKa = 3.91VPPMYY755 pKa = 11.02NHH757 pKa = 5.82TTVPAATT764 pKa = 3.93
Molecular weight: 83.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2470 |
764 |
1706 |
1235.0 |
138.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.64 ± 0.753 | 1.66 ± 0.27 |
5.385 ± 0.596 | 4.332 ± 0.666 |
6.073 ± 0.176 | 6.68 ± 0.29 |
2.146 ± 0.044 | 6.518 ± 0.161 |
4.696 ± 1.237 | 8.34 ± 0.858 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.591 ± 0.205 | 4.899 ± 0.115 |
5.223 ± 0.96 | 2.227 ± 0.219 |
5.263 ± 0.058 | 8.826 ± 0.115 |
5.628 ± 1.246 | 7.449 ± 0.519 |
1.255 ± 0.116 | 4.17 ± 0.356 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
