
Desulfuromonas thiophila
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfuromonadales; Desulfuromonadaceae; Desulfuromonas
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
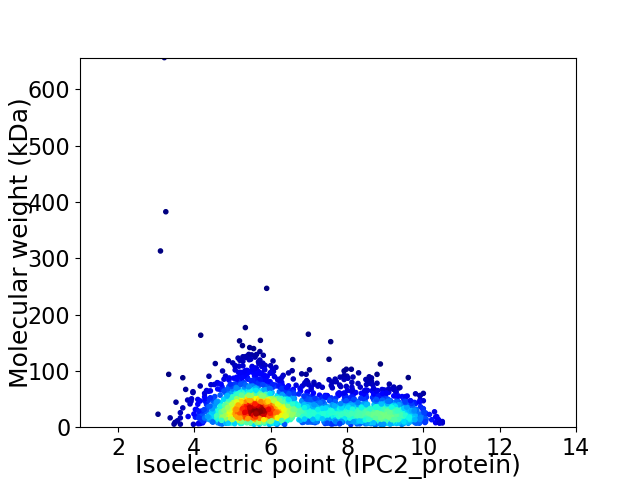
Virtual 2D-PAGE plot for 2471 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G7DPW1|A0A1G7DPW1_9DELT Uncharacterized protein OS=Desulfuromonas thiophila OX=57664 GN=SAMN05661003_1155 PE=4 SV=1
MM1 pKa = 7.07STITDD6 pKa = 3.65YY7 pKa = 11.63YY8 pKa = 10.62KK9 pKa = 10.96YY10 pKa = 10.76SEE12 pKa = 4.79LALASYY18 pKa = 11.2SDD20 pKa = 4.14LAAGMVQQDD29 pKa = 4.3YY30 pKa = 11.21IDD32 pKa = 4.77ALMNDD37 pKa = 4.43GDD39 pKa = 5.08GMSQKK44 pKa = 9.85QAEE47 pKa = 4.49TFAPNWTVLDD57 pKa = 3.85QYY59 pKa = 11.72DD60 pKa = 3.74GMVEE64 pKa = 3.69EE65 pKa = 4.86TYY67 pKa = 11.12YY68 pKa = 11.22DD69 pKa = 3.58EE70 pKa = 6.29FGDD73 pKa = 3.79EE74 pKa = 4.15QTFLNPTGLSVTLFEE89 pKa = 5.87DD90 pKa = 4.59GSGKK94 pKa = 9.82QVVAIRR100 pKa = 11.84GTEE103 pKa = 3.88PTDD106 pKa = 3.89LDD108 pKa = 5.64DD109 pKa = 4.76IVTDD113 pKa = 4.66IIDD116 pKa = 3.6IGVLGTSEE124 pKa = 4.7HH125 pKa = 5.34QAQYY129 pKa = 11.21AALSAQIQKK138 pKa = 9.86WKK140 pKa = 10.69DD141 pKa = 3.21DD142 pKa = 3.48GTLRR146 pKa = 11.84TNFSVTGHH154 pKa = 5.93SLGGFLATNLARR166 pKa = 11.84EE167 pKa = 4.21DD168 pKa = 3.87CGILKK173 pKa = 10.44
MM1 pKa = 7.07STITDD6 pKa = 3.65YY7 pKa = 11.63YY8 pKa = 10.62KK9 pKa = 10.96YY10 pKa = 10.76SEE12 pKa = 4.79LALASYY18 pKa = 11.2SDD20 pKa = 4.14LAAGMVQQDD29 pKa = 4.3YY30 pKa = 11.21IDD32 pKa = 4.77ALMNDD37 pKa = 4.43GDD39 pKa = 5.08GMSQKK44 pKa = 9.85QAEE47 pKa = 4.49TFAPNWTVLDD57 pKa = 3.85QYY59 pKa = 11.72DD60 pKa = 3.74GMVEE64 pKa = 3.69EE65 pKa = 4.86TYY67 pKa = 11.12YY68 pKa = 11.22DD69 pKa = 3.58EE70 pKa = 6.29FGDD73 pKa = 3.79EE74 pKa = 4.15QTFLNPTGLSVTLFEE89 pKa = 5.87DD90 pKa = 4.59GSGKK94 pKa = 9.82QVVAIRR100 pKa = 11.84GTEE103 pKa = 3.88PTDD106 pKa = 3.89LDD108 pKa = 5.64DD109 pKa = 4.76IVTDD113 pKa = 4.66IIDD116 pKa = 3.6IGVLGTSEE124 pKa = 4.7HH125 pKa = 5.34QAQYY129 pKa = 11.21AALSAQIQKK138 pKa = 9.86WKK140 pKa = 10.69DD141 pKa = 3.21DD142 pKa = 3.48GTLRR146 pKa = 11.84TNFSVTGHH154 pKa = 5.93SLGGFLATNLARR166 pKa = 11.84EE167 pKa = 4.21DD168 pKa = 3.87CGILKK173 pKa = 10.44
Molecular weight: 18.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6Z5H5|A0A1G6Z5H5_9DELT Diguanylate cyclase (GGDEF) domain-containing protein OS=Desulfuromonas thiophila OX=57664 GN=SAMN05661003_102306 PE=4 SV=1
MM1 pKa = 7.46QAFLHH6 pKa = 5.52NLEE9 pKa = 4.28VEE11 pKa = 4.63RR12 pKa = 11.84NLSPHH17 pKa = 5.25SCRR20 pKa = 11.84AYY22 pKa = 10.49RR23 pKa = 11.84RR24 pKa = 11.84DD25 pKa = 3.24VGAFCLFLQQQLPGRR40 pKa = 11.84PLDD43 pKa = 3.94AQLAAVDD50 pKa = 3.71KK51 pKa = 10.82ALVRR55 pKa = 11.84RR56 pKa = 11.84WLGQLGRR63 pKa = 11.84RR64 pKa = 11.84NKK66 pKa = 10.2KK67 pKa = 8.24VTLARR72 pKa = 11.84KK73 pKa = 9.27QSALRR78 pKa = 11.84TFFTFLVRR86 pKa = 11.84RR87 pKa = 11.84GLLEE91 pKa = 3.57QHH93 pKa = 7.01PLHH96 pKa = 8.3SMEE99 pKa = 4.33RR100 pKa = 11.84PRR102 pKa = 11.84LEE104 pKa = 4.07RR105 pKa = 11.84PLPRR109 pKa = 11.84LLDD112 pKa = 3.55VDD114 pKa = 5.22AVFCLLDD121 pKa = 3.64GMVPEE126 pKa = 5.26DD127 pKa = 3.22WLRR130 pKa = 11.84WRR132 pKa = 11.84DD133 pKa = 3.27RR134 pKa = 11.84ALFEE138 pKa = 4.96LLYY141 pKa = 10.85SCGLRR146 pKa = 11.84VSEE149 pKa = 5.42LVALDD154 pKa = 4.15EE155 pKa = 5.41DD156 pKa = 5.41DD157 pKa = 5.45LRR159 pKa = 11.84ADD161 pKa = 3.28QGLVCVRR168 pKa = 11.84QGKK171 pKa = 9.55GGKK174 pKa = 8.04QRR176 pKa = 11.84IVPVGQAALRR186 pKa = 11.84ALQVYY191 pKa = 8.44LQHH194 pKa = 6.95CPPQKK199 pKa = 10.04RR200 pKa = 11.84VGVALFVNQRR210 pKa = 11.84GQRR213 pKa = 11.84LSVRR217 pKa = 11.84TVQRR221 pKa = 11.84NLKK224 pKa = 9.85AALARR229 pKa = 11.84AGLPTDD235 pKa = 3.91VSPHH239 pKa = 5.28GLRR242 pKa = 11.84HH243 pKa = 5.73SFATHH248 pKa = 6.67LLDD251 pKa = 4.8GGADD255 pKa = 3.22LRR257 pKa = 11.84AIQEE261 pKa = 4.44LLGHH265 pKa = 6.96ASLATTQRR273 pKa = 11.84YY274 pKa = 5.0TQVSGARR281 pKa = 11.84LAEE284 pKa = 4.64EE285 pKa = 4.42YY286 pKa = 10.85DD287 pKa = 3.52RR288 pKa = 11.84THH290 pKa = 6.76PRR292 pKa = 11.84SKK294 pKa = 10.61KK295 pKa = 9.74PSAA298 pKa = 3.65
MM1 pKa = 7.46QAFLHH6 pKa = 5.52NLEE9 pKa = 4.28VEE11 pKa = 4.63RR12 pKa = 11.84NLSPHH17 pKa = 5.25SCRR20 pKa = 11.84AYY22 pKa = 10.49RR23 pKa = 11.84RR24 pKa = 11.84DD25 pKa = 3.24VGAFCLFLQQQLPGRR40 pKa = 11.84PLDD43 pKa = 3.94AQLAAVDD50 pKa = 3.71KK51 pKa = 10.82ALVRR55 pKa = 11.84RR56 pKa = 11.84WLGQLGRR63 pKa = 11.84RR64 pKa = 11.84NKK66 pKa = 10.2KK67 pKa = 8.24VTLARR72 pKa = 11.84KK73 pKa = 9.27QSALRR78 pKa = 11.84TFFTFLVRR86 pKa = 11.84RR87 pKa = 11.84GLLEE91 pKa = 3.57QHH93 pKa = 7.01PLHH96 pKa = 8.3SMEE99 pKa = 4.33RR100 pKa = 11.84PRR102 pKa = 11.84LEE104 pKa = 4.07RR105 pKa = 11.84PLPRR109 pKa = 11.84LLDD112 pKa = 3.55VDD114 pKa = 5.22AVFCLLDD121 pKa = 3.64GMVPEE126 pKa = 5.26DD127 pKa = 3.22WLRR130 pKa = 11.84WRR132 pKa = 11.84DD133 pKa = 3.27RR134 pKa = 11.84ALFEE138 pKa = 4.96LLYY141 pKa = 10.85SCGLRR146 pKa = 11.84VSEE149 pKa = 5.42LVALDD154 pKa = 4.15EE155 pKa = 5.41DD156 pKa = 5.41DD157 pKa = 5.45LRR159 pKa = 11.84ADD161 pKa = 3.28QGLVCVRR168 pKa = 11.84QGKK171 pKa = 9.55GGKK174 pKa = 8.04QRR176 pKa = 11.84IVPVGQAALRR186 pKa = 11.84ALQVYY191 pKa = 8.44LQHH194 pKa = 6.95CPPQKK199 pKa = 10.04RR200 pKa = 11.84VGVALFVNQRR210 pKa = 11.84GQRR213 pKa = 11.84LSVRR217 pKa = 11.84TVQRR221 pKa = 11.84NLKK224 pKa = 9.85AALARR229 pKa = 11.84AGLPTDD235 pKa = 3.91VSPHH239 pKa = 5.28GLRR242 pKa = 11.84HH243 pKa = 5.73SFATHH248 pKa = 6.67LLDD251 pKa = 4.8GGADD255 pKa = 3.22LRR257 pKa = 11.84AIQEE261 pKa = 4.44LLGHH265 pKa = 6.96ASLATTQRR273 pKa = 11.84YY274 pKa = 5.0TQVSGARR281 pKa = 11.84LAEE284 pKa = 4.64EE285 pKa = 4.42YY286 pKa = 10.85DD287 pKa = 3.52RR288 pKa = 11.84THH290 pKa = 6.76PRR292 pKa = 11.84SKK294 pKa = 10.61KK295 pKa = 9.74PSAA298 pKa = 3.65
Molecular weight: 33.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
839046 |
41 |
6445 |
339.6 |
37.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.202 ± 0.065 | 1.491 ± 0.027 |
5.369 ± 0.07 | 5.736 ± 0.045 |
3.545 ± 0.032 | 7.598 ± 0.058 |
2.103 ± 0.028 | 4.719 ± 0.04 |
2.916 ± 0.048 | 12.935 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.89 ± 0.024 | 2.653 ± 0.037 |
4.864 ± 0.047 | 5.466 ± 0.057 |
7.272 ± 0.063 | 5.256 ± 0.039 |
4.625 ± 0.056 | 6.76 ± 0.041 |
1.14 ± 0.017 | 2.461 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
