
Ruminococcus sp. AF31-8BH
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ruminococcus; unclassified Ruminococcus
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
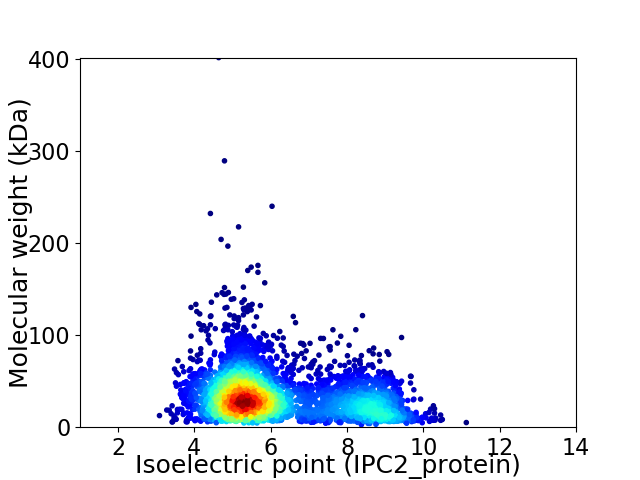
Virtual 2D-PAGE plot for 4089 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
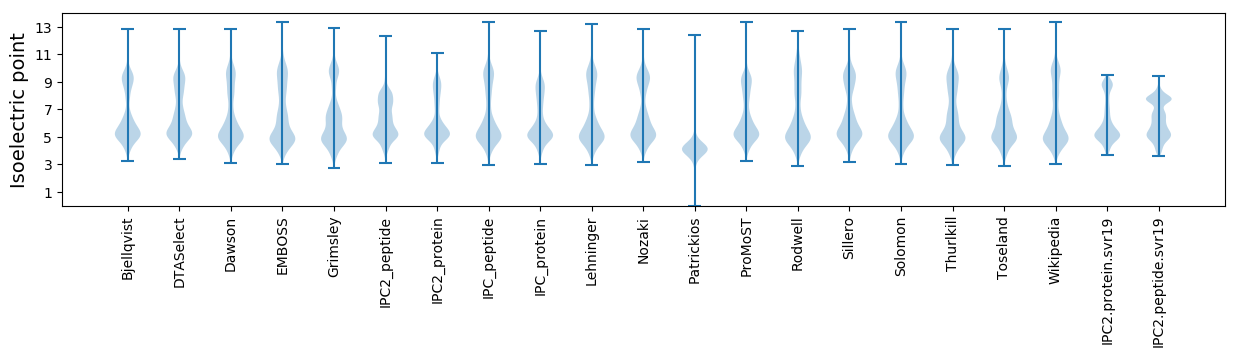
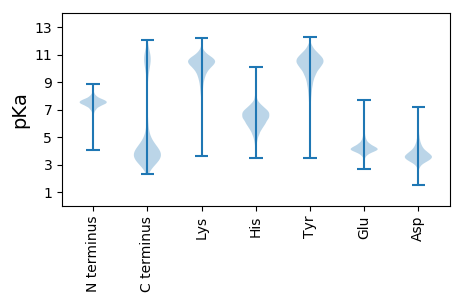
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A373NMV5|A0A373NMV5_9FIRM Hydrogenase OS=Ruminococcus sp. AF31-8BH OX=2293174 GN=DWZ38_06675 PE=3 SV=1
MM1 pKa = 7.5KK2 pKa = 10.1KK3 pKa = 9.85SKK5 pKa = 10.82AVISLLTASAMTVGLMSTGTVAFAAEE31 pKa = 4.61EE32 pKa = 4.33STSDD36 pKa = 3.7LNVMLEE42 pKa = 4.33TPVQSLDD49 pKa = 3.42PQQATDD55 pKa = 3.57GTSFEE60 pKa = 4.7VIADD64 pKa = 3.82YY65 pKa = 11.2TDD67 pKa = 4.31GLMQMDD73 pKa = 4.45SDD75 pKa = 4.26GQAVPAIAEE84 pKa = 4.38SYY86 pKa = 10.74DD87 pKa = 3.68LSDD90 pKa = 5.66DD91 pKa = 3.51GLTYY95 pKa = 10.41TFHH98 pKa = 7.78LRR100 pKa = 11.84DD101 pKa = 3.67DD102 pKa = 4.53AQWSNGDD109 pKa = 3.83PVTASDD115 pKa = 6.33FVFGWQRR122 pKa = 11.84AVDD125 pKa = 3.68PDD127 pKa = 3.78VASEE131 pKa = 3.99YY132 pKa = 10.9AYY134 pKa = 9.8MLSDD138 pKa = 3.33IGQVEE143 pKa = 4.06NAAEE147 pKa = 4.3IIAGEE152 pKa = 4.1KK153 pKa = 10.32DD154 pKa = 3.25KK155 pKa = 11.97SEE157 pKa = 4.61LGVTAVDD164 pKa = 4.36DD165 pKa = 3.65KK166 pKa = 11.29TLEE169 pKa = 4.09VKK171 pKa = 10.94LNVPVSYY178 pKa = 10.0FLSLMYY184 pKa = 10.48FPTFYY189 pKa = 10.31PVNEE193 pKa = 4.38AFFEE197 pKa = 4.54SCADD201 pKa = 3.58TFGTSPEE208 pKa = 4.23TTLSNGAFVLDD219 pKa = 4.21TYY221 pKa = 11.16EE222 pKa = 4.03PAATEE227 pKa = 3.46IHH229 pKa = 5.7LTKK232 pKa = 10.83NADD235 pKa = 3.43YY236 pKa = 11.1YY237 pKa = 11.44DD238 pKa = 4.51ADD240 pKa = 4.42NISLPGLNYY249 pKa = 10.15QVIQDD254 pKa = 3.85SQQALMSYY262 pKa = 8.01QTGDD266 pKa = 3.38LDD268 pKa = 3.73LTLVNGEE275 pKa = 4.11QVDD278 pKa = 3.99QVEE281 pKa = 4.76DD282 pKa = 3.93DD283 pKa = 4.35PEE285 pKa = 4.45FMTIGAGYY293 pKa = 9.66LWYY296 pKa = 10.58VSPNISAVPEE306 pKa = 3.96LANLNIRR313 pKa = 11.84LAMTMAIDD321 pKa = 3.95RR322 pKa = 11.84EE323 pKa = 4.59SITTDD328 pKa = 3.12VLKK331 pKa = 10.97DD332 pKa = 3.64GSLPTYY338 pKa = 9.0TAVPMQFAAGPDD350 pKa = 3.54GSDD353 pKa = 3.46FSEE356 pKa = 4.48DD357 pKa = 3.31QEE359 pKa = 4.58KK360 pKa = 10.76FSDD363 pKa = 3.61VCTYY367 pKa = 10.76DD368 pKa = 3.07AEE370 pKa = 4.33KK371 pKa = 10.75AADD374 pKa = 3.58YY375 pKa = 7.66WAKK378 pKa = 10.5GLEE381 pKa = 4.18EE382 pKa = 4.42LGEE385 pKa = 4.4TEE387 pKa = 4.75VEE389 pKa = 3.47LDD391 pKa = 4.0MIVDD395 pKa = 4.57ADD397 pKa = 4.02DD398 pKa = 5.05APQKK402 pKa = 9.76VAQVLKK408 pKa = 9.3EE409 pKa = 3.88QWEE412 pKa = 4.24TALPGLTVNLTVEE425 pKa = 4.27PKK427 pKa = 9.36KK428 pKa = 10.91QRR430 pKa = 11.84VEE432 pKa = 4.08DD433 pKa = 4.04MQNGTYY439 pKa = 9.88EE440 pKa = 4.1VALTRR445 pKa = 11.84WGPDD449 pKa = 2.99YY450 pKa = 10.98ADD452 pKa = 3.44PMTYY456 pKa = 10.43LGMWITDD463 pKa = 3.18NSNNYY468 pKa = 9.1GLWSSEE474 pKa = 4.1EE475 pKa = 3.93YY476 pKa = 10.84DD477 pKa = 4.72SIIAEE482 pKa = 4.44CTTGDD487 pKa = 3.74LCTDD491 pKa = 3.57AEE493 pKa = 4.58GRR495 pKa = 11.84WARR498 pKa = 11.84MYY500 pKa = 10.72DD501 pKa = 3.46AEE503 pKa = 4.74KK504 pKa = 10.87LVMDD508 pKa = 5.21DD509 pKa = 4.86AVIFPLYY516 pKa = 8.56TQANAEE522 pKa = 4.2MMSSSVTGVDD532 pKa = 4.28FHH534 pKa = 6.99PCALNRR540 pKa = 11.84VYY542 pKa = 11.08KK543 pKa = 10.74NAVKK547 pKa = 10.66SEE549 pKa = 3.83
MM1 pKa = 7.5KK2 pKa = 10.1KK3 pKa = 9.85SKK5 pKa = 10.82AVISLLTASAMTVGLMSTGTVAFAAEE31 pKa = 4.61EE32 pKa = 4.33STSDD36 pKa = 3.7LNVMLEE42 pKa = 4.33TPVQSLDD49 pKa = 3.42PQQATDD55 pKa = 3.57GTSFEE60 pKa = 4.7VIADD64 pKa = 3.82YY65 pKa = 11.2TDD67 pKa = 4.31GLMQMDD73 pKa = 4.45SDD75 pKa = 4.26GQAVPAIAEE84 pKa = 4.38SYY86 pKa = 10.74DD87 pKa = 3.68LSDD90 pKa = 5.66DD91 pKa = 3.51GLTYY95 pKa = 10.41TFHH98 pKa = 7.78LRR100 pKa = 11.84DD101 pKa = 3.67DD102 pKa = 4.53AQWSNGDD109 pKa = 3.83PVTASDD115 pKa = 6.33FVFGWQRR122 pKa = 11.84AVDD125 pKa = 3.68PDD127 pKa = 3.78VASEE131 pKa = 3.99YY132 pKa = 10.9AYY134 pKa = 9.8MLSDD138 pKa = 3.33IGQVEE143 pKa = 4.06NAAEE147 pKa = 4.3IIAGEE152 pKa = 4.1KK153 pKa = 10.32DD154 pKa = 3.25KK155 pKa = 11.97SEE157 pKa = 4.61LGVTAVDD164 pKa = 4.36DD165 pKa = 3.65KK166 pKa = 11.29TLEE169 pKa = 4.09VKK171 pKa = 10.94LNVPVSYY178 pKa = 10.0FLSLMYY184 pKa = 10.48FPTFYY189 pKa = 10.31PVNEE193 pKa = 4.38AFFEE197 pKa = 4.54SCADD201 pKa = 3.58TFGTSPEE208 pKa = 4.23TTLSNGAFVLDD219 pKa = 4.21TYY221 pKa = 11.16EE222 pKa = 4.03PAATEE227 pKa = 3.46IHH229 pKa = 5.7LTKK232 pKa = 10.83NADD235 pKa = 3.43YY236 pKa = 11.1YY237 pKa = 11.44DD238 pKa = 4.51ADD240 pKa = 4.42NISLPGLNYY249 pKa = 10.15QVIQDD254 pKa = 3.85SQQALMSYY262 pKa = 8.01QTGDD266 pKa = 3.38LDD268 pKa = 3.73LTLVNGEE275 pKa = 4.11QVDD278 pKa = 3.99QVEE281 pKa = 4.76DD282 pKa = 3.93DD283 pKa = 4.35PEE285 pKa = 4.45FMTIGAGYY293 pKa = 9.66LWYY296 pKa = 10.58VSPNISAVPEE306 pKa = 3.96LANLNIRR313 pKa = 11.84LAMTMAIDD321 pKa = 3.95RR322 pKa = 11.84EE323 pKa = 4.59SITTDD328 pKa = 3.12VLKK331 pKa = 10.97DD332 pKa = 3.64GSLPTYY338 pKa = 9.0TAVPMQFAAGPDD350 pKa = 3.54GSDD353 pKa = 3.46FSEE356 pKa = 4.48DD357 pKa = 3.31QEE359 pKa = 4.58KK360 pKa = 10.76FSDD363 pKa = 3.61VCTYY367 pKa = 10.76DD368 pKa = 3.07AEE370 pKa = 4.33KK371 pKa = 10.75AADD374 pKa = 3.58YY375 pKa = 7.66WAKK378 pKa = 10.5GLEE381 pKa = 4.18EE382 pKa = 4.42LGEE385 pKa = 4.4TEE387 pKa = 4.75VEE389 pKa = 3.47LDD391 pKa = 4.0MIVDD395 pKa = 4.57ADD397 pKa = 4.02DD398 pKa = 5.05APQKK402 pKa = 9.76VAQVLKK408 pKa = 9.3EE409 pKa = 3.88QWEE412 pKa = 4.24TALPGLTVNLTVEE425 pKa = 4.27PKK427 pKa = 9.36KK428 pKa = 10.91QRR430 pKa = 11.84VEE432 pKa = 4.08DD433 pKa = 4.04MQNGTYY439 pKa = 9.88EE440 pKa = 4.1VALTRR445 pKa = 11.84WGPDD449 pKa = 2.99YY450 pKa = 10.98ADD452 pKa = 3.44PMTYY456 pKa = 10.43LGMWITDD463 pKa = 3.18NSNNYY468 pKa = 9.1GLWSSEE474 pKa = 4.1EE475 pKa = 3.93YY476 pKa = 10.84DD477 pKa = 4.72SIIAEE482 pKa = 4.44CTTGDD487 pKa = 3.74LCTDD491 pKa = 3.57AEE493 pKa = 4.58GRR495 pKa = 11.84WARR498 pKa = 11.84MYY500 pKa = 10.72DD501 pKa = 3.46AEE503 pKa = 4.74KK504 pKa = 10.87LVMDD508 pKa = 5.21DD509 pKa = 4.86AVIFPLYY516 pKa = 8.56TQANAEE522 pKa = 4.2MMSSSVTGVDD532 pKa = 4.28FHH534 pKa = 6.99PCALNRR540 pKa = 11.84VYY542 pKa = 11.08KK543 pKa = 10.74NAVKK547 pKa = 10.66SEE549 pKa = 3.83
Molecular weight: 60.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A373NU95|A0A373NU95_9FIRM Peptidyl-tRNA hydrolase OS=Ruminococcus sp. AF31-8BH OX=2293174 GN=pth PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 8.28QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 8.28QLSAA44 pKa = 3.9
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1301907 |
24 |
3759 |
318.4 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.417 ± 0.043 | 1.555 ± 0.016 |
5.629 ± 0.035 | 7.691 ± 0.045 |
4.113 ± 0.028 | 7.031 ± 0.04 |
1.784 ± 0.018 | 7.037 ± 0.031 |
6.949 ± 0.036 | 8.936 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.154 ± 0.02 | 4.416 ± 0.028 |
3.308 ± 0.026 | 3.507 ± 0.023 |
4.325 ± 0.034 | 5.785 ± 0.033 |
5.412 ± 0.034 | 6.73 ± 0.03 |
0.998 ± 0.012 | 4.223 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
