
Beluga whale alphaherpesvirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Varicellovirus; Monodontid alphaherpesvirus 1
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
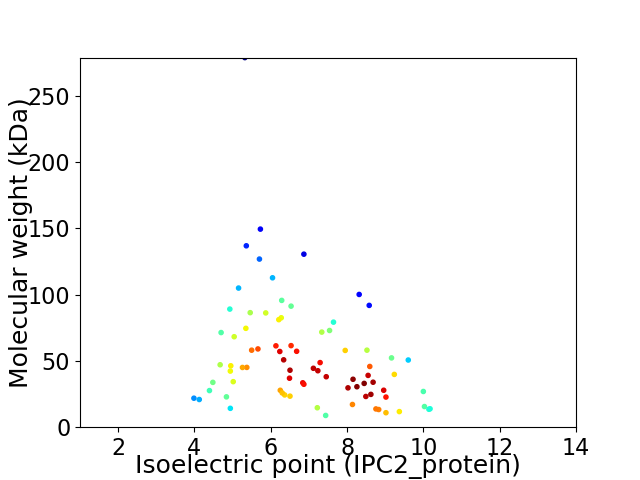
Virtual 2D-PAGE plot for 80 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
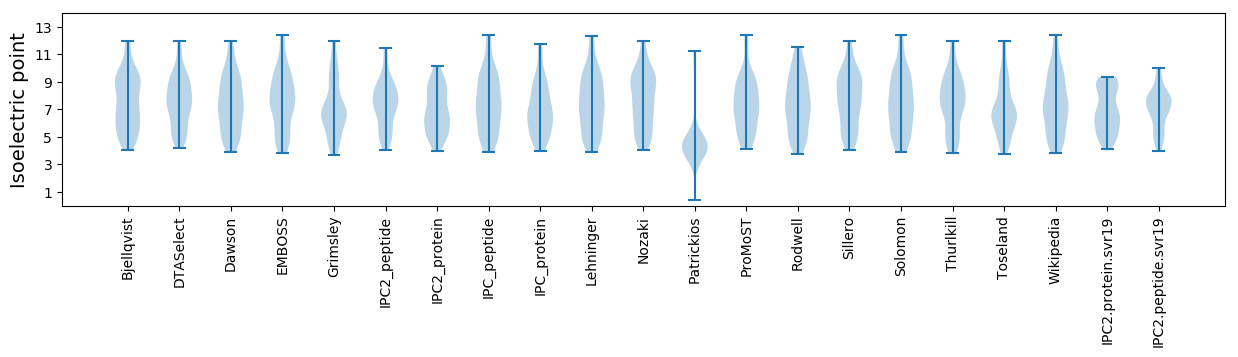
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A286MM97|A0A286MM97_9ALPH Protein Mo1 OS=Beluga whale alphaherpesvirus 1 OX=1434720 GN=Mo1 PE=4 SV=1
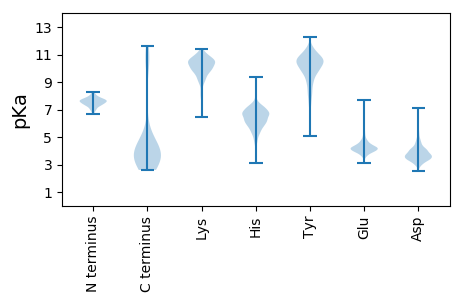
MM1 pKa = 7.49AAFVSDD7 pKa = 3.41RR8 pKa = 11.84WFDD11 pKa = 3.64AYY13 pKa = 10.59MGDD16 pKa = 3.69PWSEE20 pKa = 3.71PVYY23 pKa = 11.09VSLTTPLSEE32 pKa = 4.18SAPRR36 pKa = 11.84GTPRR40 pKa = 11.84RR41 pKa = 11.84APSTGSPPPSPDD53 pKa = 3.35PEE55 pKa = 4.19RR56 pKa = 11.84APPLPPDD63 pKa = 4.48PLPPVPDD70 pKa = 4.3SPPPPYY76 pKa = 9.65PAAEE80 pKa = 4.25TPPSFPLADD89 pKa = 3.69PARR92 pKa = 11.84GDD94 pKa = 3.77EE95 pKa = 4.47APGAPEE101 pKa = 4.29DD102 pKa = 3.92ARR104 pKa = 11.84ITATSVEE111 pKa = 4.25NPSYY115 pKa = 10.67LGPLLLPAGLPDD127 pKa = 4.68RR128 pKa = 11.84PPAYY132 pKa = 8.12DD133 pKa = 3.64TLSVALPPPSYY144 pKa = 10.2EE145 pKa = 3.68WAIMDD150 pKa = 5.33DD151 pKa = 4.1LLASIAYY158 pKa = 7.5PRR160 pKa = 11.84LGEE163 pKa = 4.19KK164 pKa = 9.69EE165 pKa = 4.16CCNVVGLAALLILIIILFGVFAYY188 pKa = 9.71ILAGSGGGMSKK199 pKa = 10.77
MM1 pKa = 7.49AAFVSDD7 pKa = 3.41RR8 pKa = 11.84WFDD11 pKa = 3.64AYY13 pKa = 10.59MGDD16 pKa = 3.69PWSEE20 pKa = 3.71PVYY23 pKa = 11.09VSLTTPLSEE32 pKa = 4.18SAPRR36 pKa = 11.84GTPRR40 pKa = 11.84RR41 pKa = 11.84APSTGSPPPSPDD53 pKa = 3.35PEE55 pKa = 4.19RR56 pKa = 11.84APPLPPDD63 pKa = 4.48PLPPVPDD70 pKa = 4.3SPPPPYY76 pKa = 9.65PAAEE80 pKa = 4.25TPPSFPLADD89 pKa = 3.69PARR92 pKa = 11.84GDD94 pKa = 3.77EE95 pKa = 4.47APGAPEE101 pKa = 4.29DD102 pKa = 3.92ARR104 pKa = 11.84ITATSVEE111 pKa = 4.25NPSYY115 pKa = 10.67LGPLLLPAGLPDD127 pKa = 4.68RR128 pKa = 11.84PPAYY132 pKa = 8.12DD133 pKa = 3.64TLSVALPPPSYY144 pKa = 10.2EE145 pKa = 3.68WAIMDD150 pKa = 5.33DD151 pKa = 4.1LLASIAYY158 pKa = 7.5PRR160 pKa = 11.84LGEE163 pKa = 4.19KK164 pKa = 9.69EE165 pKa = 4.16CCNVVGLAALLILIIILFGVFAYY188 pKa = 9.71ILAGSGGGMSKK199 pKa = 10.77
Molecular weight: 20.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A286MM96|A0A286MM96_9ALPH Protein Mo7 OS=Beluga whale alphaherpesvirus 1 OX=1434720 GN=Mo7 PE=4 SV=1
MM1 pKa = 7.79DD2 pKa = 5.43RR3 pKa = 11.84GAAGLGRR10 pKa = 11.84AACALQVWLALGLARR25 pKa = 11.84GVGAHH30 pKa = 6.48TDD32 pKa = 3.57DD33 pKa = 5.2EE34 pKa = 5.64IDD36 pKa = 3.45VSEE39 pKa = 4.57YY40 pKa = 10.33PVYY43 pKa = 10.65NCSLVAVRR51 pKa = 11.84NFAGWFVPPAYY62 pKa = 10.06AVICLVGVFGNLLVLAMFVTYY83 pKa = 10.53AKK85 pKa = 10.11SRR87 pKa = 11.84CLTDD91 pKa = 3.65VFIANMALADD101 pKa = 4.29LLLVLTLPFWAVQHH115 pKa = 6.91ALDD118 pKa = 4.71DD119 pKa = 4.66WIFSNIACKK128 pKa = 7.95VTRR131 pKa = 11.84GIYY134 pKa = 10.19AINFNCGMLLLTCIGVDD151 pKa = 3.31RR152 pKa = 11.84YY153 pKa = 10.33IAIVRR158 pKa = 11.84ASTAARR164 pKa = 11.84LRR166 pKa = 11.84DD167 pKa = 3.64RR168 pKa = 11.84LRR170 pKa = 11.84ARR172 pKa = 11.84SGLICVGVWAVALATASGTFVFTQKK197 pKa = 10.63YY198 pKa = 8.63HH199 pKa = 4.82VTNSTHH205 pKa = 5.47VCDD208 pKa = 4.86ARR210 pKa = 11.84FDD212 pKa = 4.14TTSSAPLVWRR222 pKa = 11.84RR223 pKa = 11.84LMLSVQLASGFFFPVVCMGVFYY245 pKa = 9.88FLIIRR250 pKa = 11.84RR251 pKa = 11.84LLRR254 pKa = 11.84SINARR259 pKa = 11.84KK260 pKa = 8.06HH261 pKa = 3.61TAIRR265 pKa = 11.84VSLAMVLVFFVCQLPHH281 pKa = 6.68NSVLLMTAVNSHH293 pKa = 5.18GMVVHH298 pKa = 6.35NRR300 pKa = 11.84TDD302 pKa = 3.09NVTEE306 pKa = 3.99IWCPIMRR313 pKa = 11.84NLRR316 pKa = 11.84YY317 pKa = 9.28AQRR320 pKa = 11.84ATEE323 pKa = 4.14VLAFLHH329 pKa = 6.55GSVNPVLYY337 pKa = 10.77AFLGKK342 pKa = 10.21KK343 pKa = 9.65FRR345 pKa = 11.84EE346 pKa = 3.88QFLRR350 pKa = 11.84MVRR353 pKa = 11.84RR354 pKa = 11.84RR355 pKa = 11.84GAAPAMRR362 pKa = 11.84ISSPVKK368 pKa = 9.67RR369 pKa = 11.84PSISPPPARR378 pKa = 11.84PLLGAVCGPPRR389 pKa = 11.84SPPPKK394 pKa = 9.39TVFTFPSDD402 pKa = 3.55GFSGPVFPLVVAFPAALPAPPHH424 pKa = 5.38SLGWVSSPEE433 pKa = 3.83RR434 pKa = 11.84PGRR437 pKa = 11.84VARR440 pKa = 11.84PLDD443 pKa = 3.66SAPRR447 pKa = 11.84RR448 pKa = 11.84ARR450 pKa = 11.84TRR452 pKa = 11.84GTPAAHH458 pKa = 5.8VEE460 pKa = 4.38KK461 pKa = 11.09VV462 pKa = 2.98
MM1 pKa = 7.79DD2 pKa = 5.43RR3 pKa = 11.84GAAGLGRR10 pKa = 11.84AACALQVWLALGLARR25 pKa = 11.84GVGAHH30 pKa = 6.48TDD32 pKa = 3.57DD33 pKa = 5.2EE34 pKa = 5.64IDD36 pKa = 3.45VSEE39 pKa = 4.57YY40 pKa = 10.33PVYY43 pKa = 10.65NCSLVAVRR51 pKa = 11.84NFAGWFVPPAYY62 pKa = 10.06AVICLVGVFGNLLVLAMFVTYY83 pKa = 10.53AKK85 pKa = 10.11SRR87 pKa = 11.84CLTDD91 pKa = 3.65VFIANMALADD101 pKa = 4.29LLLVLTLPFWAVQHH115 pKa = 6.91ALDD118 pKa = 4.71DD119 pKa = 4.66WIFSNIACKK128 pKa = 7.95VTRR131 pKa = 11.84GIYY134 pKa = 10.19AINFNCGMLLLTCIGVDD151 pKa = 3.31RR152 pKa = 11.84YY153 pKa = 10.33IAIVRR158 pKa = 11.84ASTAARR164 pKa = 11.84LRR166 pKa = 11.84DD167 pKa = 3.64RR168 pKa = 11.84LRR170 pKa = 11.84ARR172 pKa = 11.84SGLICVGVWAVALATASGTFVFTQKK197 pKa = 10.63YY198 pKa = 8.63HH199 pKa = 4.82VTNSTHH205 pKa = 5.47VCDD208 pKa = 4.86ARR210 pKa = 11.84FDD212 pKa = 4.14TTSSAPLVWRR222 pKa = 11.84RR223 pKa = 11.84LMLSVQLASGFFFPVVCMGVFYY245 pKa = 9.88FLIIRR250 pKa = 11.84RR251 pKa = 11.84LLRR254 pKa = 11.84SINARR259 pKa = 11.84KK260 pKa = 8.06HH261 pKa = 3.61TAIRR265 pKa = 11.84VSLAMVLVFFVCQLPHH281 pKa = 6.68NSVLLMTAVNSHH293 pKa = 5.18GMVVHH298 pKa = 6.35NRR300 pKa = 11.84TDD302 pKa = 3.09NVTEE306 pKa = 3.99IWCPIMRR313 pKa = 11.84NLRR316 pKa = 11.84YY317 pKa = 9.28AQRR320 pKa = 11.84ATEE323 pKa = 4.14VLAFLHH329 pKa = 6.55GSVNPVLYY337 pKa = 10.77AFLGKK342 pKa = 10.21KK343 pKa = 9.65FRR345 pKa = 11.84EE346 pKa = 3.88QFLRR350 pKa = 11.84MVRR353 pKa = 11.84RR354 pKa = 11.84RR355 pKa = 11.84GAAPAMRR362 pKa = 11.84ISSPVKK368 pKa = 9.67RR369 pKa = 11.84PSISPPPARR378 pKa = 11.84PLLGAVCGPPRR389 pKa = 11.84SPPPKK394 pKa = 9.39TVFTFPSDD402 pKa = 3.55GFSGPVFPLVVAFPAALPAPPHH424 pKa = 5.38SLGWVSSPEE433 pKa = 3.83RR434 pKa = 11.84PGRR437 pKa = 11.84VARR440 pKa = 11.84PLDD443 pKa = 3.66SAPRR447 pKa = 11.84RR448 pKa = 11.84ARR450 pKa = 11.84TRR452 pKa = 11.84GTPAAHH458 pKa = 5.8VEE460 pKa = 4.38KK461 pKa = 11.09VV462 pKa = 2.98
Molecular weight: 50.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
39899 |
79 |
2627 |
498.7 |
53.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.316 ± 0.466 | 1.857 ± 0.101 |
5.699 ± 0.148 | 5.379 ± 0.191 |
3.406 ± 0.184 | 8.025 ± 0.205 |
2.095 ± 0.099 | 2.436 ± 0.133 |
1.556 ± 0.138 | 9.592 ± 0.28 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.511 ± 0.098 | 1.957 ± 0.158 |
8.346 ± 0.377 | 2.045 ± 0.122 |
9.105 ± 0.242 | 5.755 ± 0.192 |
4.827 ± 0.159 | 7.308 ± 0.199 |
1.14 ± 0.066 | 2.642 ± 0.132 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
