
Eastern grey kangaroopox virus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Chitovirales; Poxviridae; Chordopoxvirinae; Macropopoxvirus; Eastern kangaroopox virus
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
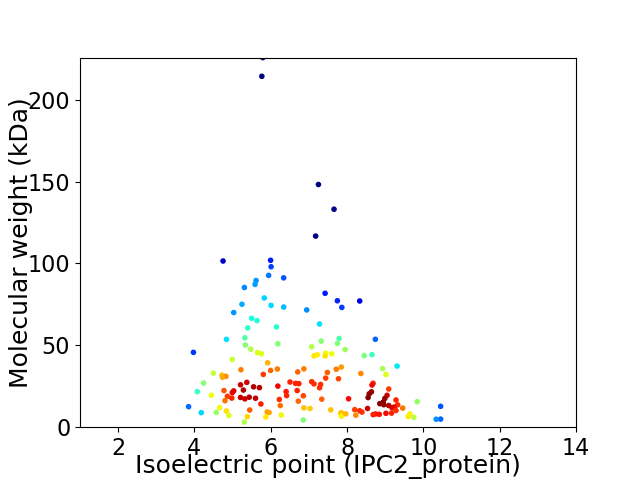
Virtual 2D-PAGE plot for 162 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2C9DSW8|A0A2C9DSW8_9POXV Uncharacterized protein OS=Eastern grey kangaroopox virus OX=2042482 GN=EKPV-NSW-ORF010 PE=4 SV=1
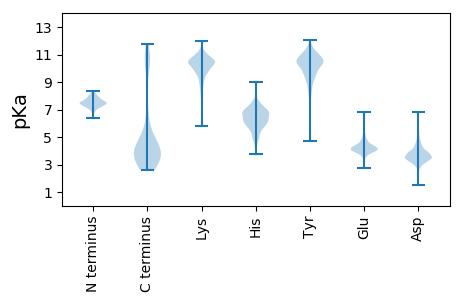
MM1 pKa = 7.34FGIRR5 pKa = 11.84NTRR8 pKa = 11.84FCLSCEE14 pKa = 4.31TTNHH18 pKa = 4.37VNIYY22 pKa = 9.63SVSAEE27 pKa = 4.02VCYY30 pKa = 10.57LPPKK34 pKa = 10.32FLEE37 pKa = 4.23TLDD40 pKa = 4.56LCVQEE45 pKa = 5.22SDD47 pKa = 4.68CGCCYY52 pKa = 10.44KK53 pKa = 10.51SEE55 pKa = 4.42AFLNTPKK62 pKa = 9.88CTLPPHH68 pKa = 5.99ITCAFCDD75 pKa = 3.79TYY77 pKa = 11.55KK78 pKa = 10.81RR79 pKa = 11.84LHH81 pKa = 6.53KK82 pKa = 9.9PDD84 pKa = 3.75TGTSFQRR91 pKa = 11.84SVEE94 pKa = 4.1DD95 pKa = 4.0DD96 pKa = 3.73VQGTICGMEE105 pKa = 4.13EE106 pKa = 4.3DD107 pKa = 4.7DD108 pKa = 5.63VSILEE113 pKa = 4.07PAAIEE118 pKa = 4.25AEE120 pKa = 4.32TTEE123 pKa = 4.4EE124 pKa = 3.94EE125 pKa = 4.53VEE127 pKa = 4.1PEE129 pKa = 3.85EE130 pKa = 4.29TEE132 pKa = 4.26EE133 pKa = 3.96EE134 pKa = 4.46VEE136 pKa = 4.28PEE138 pKa = 3.87EE139 pKa = 4.41TEE141 pKa = 4.12AGAEE145 pKa = 3.95ALEE148 pKa = 4.34DD149 pKa = 4.02LLRR152 pKa = 11.84CDD154 pKa = 3.96EE155 pKa = 4.47SDD157 pKa = 3.99LIEE160 pKa = 5.22DD161 pKa = 3.71EE162 pKa = 5.28GVTFPHH168 pKa = 7.3DD169 pKa = 4.0KK170 pKa = 11.01KK171 pKa = 10.85NLPNMLYY178 pKa = 10.43ADD180 pKa = 4.26PEE182 pKa = 4.06AALMSEE188 pKa = 4.5EE189 pKa = 4.61EE190 pKa = 4.3YY191 pKa = 10.56MSFVNAEE198 pKa = 3.66LDD200 pKa = 3.76YY201 pKa = 10.61YY202 pKa = 11.51LEE204 pKa = 4.33VADD207 pKa = 5.55AEE209 pKa = 4.61VDD211 pKa = 3.9TEE213 pKa = 3.91WRR215 pKa = 11.84RR216 pKa = 11.84DD217 pKa = 3.62LEE219 pKa = 4.21RR220 pKa = 11.84LPGPVPEE227 pKa = 4.54FDD229 pKa = 4.9AVRR232 pKa = 11.84GWDD235 pKa = 3.57VDD237 pKa = 3.82PAGVWVAAGDD247 pKa = 3.94VVDD250 pKa = 5.97AEE252 pKa = 4.56EE253 pKa = 4.98DD254 pKa = 3.34WGQGPQAIWDD264 pKa = 3.89HH265 pKa = 6.32EE266 pKa = 4.52DD267 pKa = 3.06VGRR270 pKa = 11.84EE271 pKa = 3.98QEE273 pKa = 4.25WVPEE277 pKa = 4.02AGVVDD282 pKa = 4.18RR283 pKa = 11.84RR284 pKa = 11.84DD285 pKa = 3.56EE286 pKa = 3.86EE287 pKa = 4.24HH288 pKa = 7.21LIDD291 pKa = 5.14NRR293 pKa = 11.84YY294 pKa = 9.32DD295 pKa = 3.42HH296 pKa = 6.08YY297 pKa = 11.46AIWRR301 pKa = 11.84SIFDD305 pKa = 3.77EE306 pKa = 4.5RR307 pKa = 11.84QGGAPNEE314 pKa = 4.41DD315 pKa = 3.58EE316 pKa = 6.01DD317 pKa = 5.21EE318 pKa = 5.13IGDD321 pKa = 3.97SSFLCEE327 pKa = 5.29CDD329 pKa = 2.94TCEE332 pKa = 4.36YY333 pKa = 10.52ISDD336 pKa = 4.07PVQGHH341 pKa = 7.16LIIADD346 pKa = 3.44NDD348 pKa = 3.96GVVSVDD354 pKa = 3.54VLNGSHH360 pKa = 6.6TDD362 pKa = 3.61AVPSLCLAAYY372 pKa = 9.25LHH374 pKa = 6.33SCRR377 pKa = 11.84SGEE380 pKa = 3.92IEE382 pKa = 4.17FVKK385 pKa = 10.41HH386 pKa = 5.79YY387 pKa = 10.22IVQEE391 pKa = 4.04FFGGPPTSSIALRR404 pKa = 11.84RR405 pKa = 11.84CNN407 pKa = 3.5
MM1 pKa = 7.34FGIRR5 pKa = 11.84NTRR8 pKa = 11.84FCLSCEE14 pKa = 4.31TTNHH18 pKa = 4.37VNIYY22 pKa = 9.63SVSAEE27 pKa = 4.02VCYY30 pKa = 10.57LPPKK34 pKa = 10.32FLEE37 pKa = 4.23TLDD40 pKa = 4.56LCVQEE45 pKa = 5.22SDD47 pKa = 4.68CGCCYY52 pKa = 10.44KK53 pKa = 10.51SEE55 pKa = 4.42AFLNTPKK62 pKa = 9.88CTLPPHH68 pKa = 5.99ITCAFCDD75 pKa = 3.79TYY77 pKa = 11.55KK78 pKa = 10.81RR79 pKa = 11.84LHH81 pKa = 6.53KK82 pKa = 9.9PDD84 pKa = 3.75TGTSFQRR91 pKa = 11.84SVEE94 pKa = 4.1DD95 pKa = 4.0DD96 pKa = 3.73VQGTICGMEE105 pKa = 4.13EE106 pKa = 4.3DD107 pKa = 4.7DD108 pKa = 5.63VSILEE113 pKa = 4.07PAAIEE118 pKa = 4.25AEE120 pKa = 4.32TTEE123 pKa = 4.4EE124 pKa = 3.94EE125 pKa = 4.53VEE127 pKa = 4.1PEE129 pKa = 3.85EE130 pKa = 4.29TEE132 pKa = 4.26EE133 pKa = 3.96EE134 pKa = 4.46VEE136 pKa = 4.28PEE138 pKa = 3.87EE139 pKa = 4.41TEE141 pKa = 4.12AGAEE145 pKa = 3.95ALEE148 pKa = 4.34DD149 pKa = 4.02LLRR152 pKa = 11.84CDD154 pKa = 3.96EE155 pKa = 4.47SDD157 pKa = 3.99LIEE160 pKa = 5.22DD161 pKa = 3.71EE162 pKa = 5.28GVTFPHH168 pKa = 7.3DD169 pKa = 4.0KK170 pKa = 11.01KK171 pKa = 10.85NLPNMLYY178 pKa = 10.43ADD180 pKa = 4.26PEE182 pKa = 4.06AALMSEE188 pKa = 4.5EE189 pKa = 4.61EE190 pKa = 4.3YY191 pKa = 10.56MSFVNAEE198 pKa = 3.66LDD200 pKa = 3.76YY201 pKa = 10.61YY202 pKa = 11.51LEE204 pKa = 4.33VADD207 pKa = 5.55AEE209 pKa = 4.61VDD211 pKa = 3.9TEE213 pKa = 3.91WRR215 pKa = 11.84RR216 pKa = 11.84DD217 pKa = 3.62LEE219 pKa = 4.21RR220 pKa = 11.84LPGPVPEE227 pKa = 4.54FDD229 pKa = 4.9AVRR232 pKa = 11.84GWDD235 pKa = 3.57VDD237 pKa = 3.82PAGVWVAAGDD247 pKa = 3.94VVDD250 pKa = 5.97AEE252 pKa = 4.56EE253 pKa = 4.98DD254 pKa = 3.34WGQGPQAIWDD264 pKa = 3.89HH265 pKa = 6.32EE266 pKa = 4.52DD267 pKa = 3.06VGRR270 pKa = 11.84EE271 pKa = 3.98QEE273 pKa = 4.25WVPEE277 pKa = 4.02AGVVDD282 pKa = 4.18RR283 pKa = 11.84RR284 pKa = 11.84DD285 pKa = 3.56EE286 pKa = 3.86EE287 pKa = 4.24HH288 pKa = 7.21LIDD291 pKa = 5.14NRR293 pKa = 11.84YY294 pKa = 9.32DD295 pKa = 3.42HH296 pKa = 6.08YY297 pKa = 11.46AIWRR301 pKa = 11.84SIFDD305 pKa = 3.77EE306 pKa = 4.5RR307 pKa = 11.84QGGAPNEE314 pKa = 4.41DD315 pKa = 3.58EE316 pKa = 6.01DD317 pKa = 5.21EE318 pKa = 5.13IGDD321 pKa = 3.97SSFLCEE327 pKa = 5.29CDD329 pKa = 2.94TCEE332 pKa = 4.36YY333 pKa = 10.52ISDD336 pKa = 4.07PVQGHH341 pKa = 7.16LIIADD346 pKa = 3.44NDD348 pKa = 3.96GVVSVDD354 pKa = 3.54VLNGSHH360 pKa = 6.6TDD362 pKa = 3.61AVPSLCLAAYY372 pKa = 9.25LHH374 pKa = 6.33SCRR377 pKa = 11.84SGEE380 pKa = 3.92IEE382 pKa = 4.17FVKK385 pKa = 10.41HH386 pKa = 5.79YY387 pKa = 10.22IVQEE391 pKa = 4.04FFGGPPTSSIALRR404 pKa = 11.84RR405 pKa = 11.84CNN407 pKa = 3.5
Molecular weight: 45.7 kDa
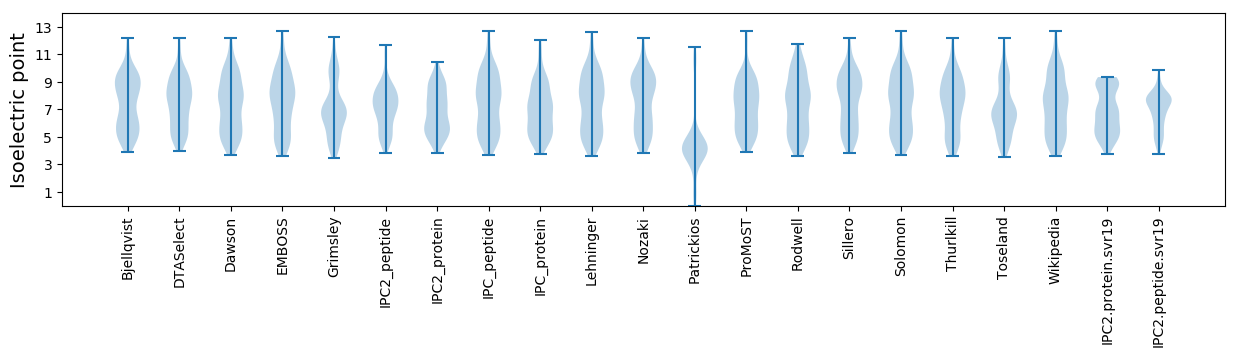
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2C9DSY8|A0A2C9DSY8_9POXV S-S bond formation pathway protein substrate OS=Eastern grey kangaroopox virus OX=2042482 GN=EKPV-NSW-ORF038 PE=4 SV=1
MM1 pKa = 7.09RR2 pKa = 11.84QLWRR6 pKa = 11.84LRR8 pKa = 11.84VLGAKK13 pKa = 9.92KK14 pKa = 10.35KK15 pKa = 10.99GGDD18 pKa = 4.05GDD20 pKa = 4.71RR21 pKa = 11.84GDD23 pKa = 4.52CGGGDD28 pKa = 3.56SSDD31 pKa = 3.51CLIYY35 pKa = 10.53SIYY38 pKa = 10.12RR39 pKa = 11.84VCNLPVRR46 pKa = 11.84QPVRR50 pKa = 11.84SGRR53 pKa = 11.84LARR56 pKa = 11.84RR57 pKa = 11.84WW58 pKa = 3.32
MM1 pKa = 7.09RR2 pKa = 11.84QLWRR6 pKa = 11.84LRR8 pKa = 11.84VLGAKK13 pKa = 9.92KK14 pKa = 10.35KK15 pKa = 10.99GGDD18 pKa = 4.05GDD20 pKa = 4.71RR21 pKa = 11.84GDD23 pKa = 4.52CGGGDD28 pKa = 3.56SSDD31 pKa = 3.51CLIYY35 pKa = 10.53SIYY38 pKa = 10.12RR39 pKa = 11.84VCNLPVRR46 pKa = 11.84QPVRR50 pKa = 11.84SGRR53 pKa = 11.84LARR56 pKa = 11.84RR57 pKa = 11.84WW58 pKa = 3.32
Molecular weight: 6.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
51353 |
32 |
2030 |
317.0 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.942 ± 0.259 | 2.206 ± 0.126 |
5.624 ± 0.116 | 6.368 ± 0.143 |
4.475 ± 0.148 | 5.419 ± 0.229 |
2.255 ± 0.097 | 5.388 ± 0.222 |
4.011 ± 0.181 | 9.762 ± 0.183 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.454 ± 0.08 | 4.019 ± 0.164 |
4.539 ± 0.213 | 2.019 ± 0.07 |
7.55 ± 0.19 | 8.543 ± 0.266 |
5.396 ± 0.129 | 8.136 ± 0.146 |
0.61 ± 0.043 | 4.284 ± 0.125 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
