
Salinicoccus alkaliphilus DSM 16010
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Staphylococcaceae; Salinicoccus; Salinicoccus alkaliphilus
Average proteome isoelectric point is 5.75
Get precalculated fractions of proteins
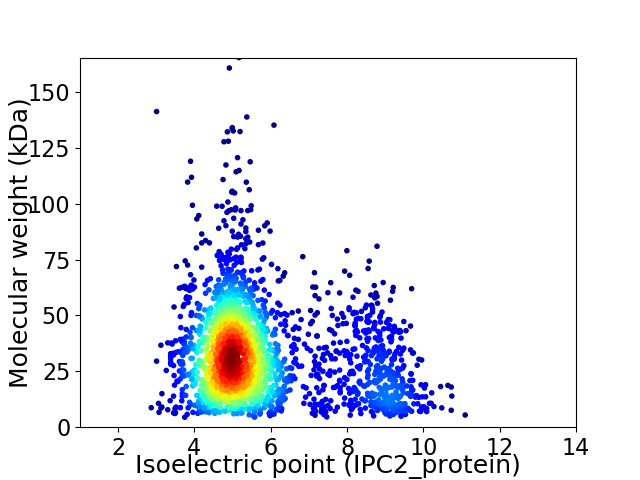
Virtual 2D-PAGE plot for 2512 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M7HGP6|A0A1M7HGP6_9STAP Uncharacterized protein OS=Salinicoccus alkaliphilus DSM 16010 OX=1123231 GN=SAMN02745189_01859 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 10.2KK3 pKa = 10.49YY4 pKa = 10.75LILVLSLVFVLAACTDD20 pKa = 3.74DD21 pKa = 5.31GEE23 pKa = 4.36VDD25 pKa = 3.25EE26 pKa = 4.88ATEE29 pKa = 4.1EE30 pKa = 4.21EE31 pKa = 4.64ATGEE35 pKa = 4.04DD36 pKa = 3.67AAEE39 pKa = 4.36EE40 pKa = 4.8GSTGGSEE47 pKa = 4.14GTINMSLLATPNSLDD62 pKa = 3.18PHH64 pKa = 6.75AANDD68 pKa = 3.63QPSNHH73 pKa = 5.51VNVNIYY79 pKa = 9.9EE80 pKa = 4.19RR81 pKa = 11.84LVEE84 pKa = 4.3FTPEE88 pKa = 3.95LEE90 pKa = 4.45LEE92 pKa = 4.58PGLAEE97 pKa = 4.44SYY99 pKa = 10.5EE100 pKa = 4.15QVEE103 pKa = 4.35DD104 pKa = 3.98TTWEE108 pKa = 3.8FSIRR112 pKa = 11.84EE113 pKa = 4.16GVSFHH118 pKa = 7.68DD119 pKa = 4.24GEE121 pKa = 4.34EE122 pKa = 4.21LNAEE126 pKa = 4.0AVKK129 pKa = 11.03ANLDD133 pKa = 3.53RR134 pKa = 11.84VTDD137 pKa = 3.83EE138 pKa = 5.32DD139 pKa = 3.79VGSPVAFLFEE149 pKa = 5.19LIEE152 pKa = 4.12EE153 pKa = 4.45VEE155 pKa = 4.51VIDD158 pKa = 5.63DD159 pKa = 4.04YY160 pKa = 11.32TVHH163 pKa = 6.38IHH165 pKa = 5.78TSSPFAALPSHH176 pKa = 6.99LAHH179 pKa = 7.24PAGGMISPAVIEE191 pKa = 3.89EE192 pKa = 4.75DD193 pKa = 3.84YY194 pKa = 11.8AEE196 pKa = 5.04DD197 pKa = 3.71EE198 pKa = 4.34QLTAVHH204 pKa = 5.94QNPVGTGAYY213 pKa = 8.83QFEE216 pKa = 4.91EE217 pKa = 4.42ISEE220 pKa = 4.18GDD222 pKa = 3.73YY223 pKa = 10.0VTLVKK228 pKa = 10.87NEE230 pKa = 4.25EE231 pKa = 4.22YY232 pKa = 9.71WGEE235 pKa = 3.93EE236 pKa = 3.95AEE238 pKa = 4.29SDD240 pKa = 3.85VFTFNAVPEE249 pKa = 4.44DD250 pKa = 3.42ATRR253 pKa = 11.84IAEE256 pKa = 4.17LQTRR260 pKa = 11.84HH261 pKa = 6.38ADD263 pKa = 3.99LIFPLDD269 pKa = 4.02PNDD272 pKa = 3.78FEE274 pKa = 5.63QINSDD279 pKa = 2.94EE280 pKa = 4.17GTTVNEE286 pKa = 4.3TEE288 pKa = 4.51SVRR291 pKa = 11.84MEE293 pKa = 3.64YY294 pKa = 10.69VGFNVEE300 pKa = 4.59VEE302 pKa = 4.31PFDD305 pKa = 5.19DD306 pKa = 3.96PVVRR310 pKa = 11.84QAIAHH315 pKa = 6.97AIDD318 pKa = 4.37KK319 pKa = 10.62EE320 pKa = 4.32DD321 pKa = 4.4IINIMLEE328 pKa = 4.07GKK330 pKa = 10.29AVVADD335 pKa = 4.11TLLSPAVFGHH345 pKa = 6.97AEE347 pKa = 4.49DD348 pKa = 4.19IDD350 pKa = 4.83GIGYY354 pKa = 9.75DD355 pKa = 4.23LEE357 pKa = 4.56TAQEE361 pKa = 4.07LLDD364 pKa = 4.31DD365 pKa = 4.31NGYY368 pKa = 10.76EE369 pKa = 4.06EE370 pKa = 5.3GFSAEE375 pKa = 4.02IVVQDD380 pKa = 3.88RR381 pKa = 11.84TAADD385 pKa = 2.99IATYY389 pKa = 9.92IQEE392 pKa = 4.21QLQEE396 pKa = 4.19LNIDD400 pKa = 3.72LEE402 pKa = 4.74IYY404 pKa = 10.56QIDD407 pKa = 3.51PGAYY411 pKa = 8.92LDD413 pKa = 4.05YY414 pKa = 10.83VGSGSHH420 pKa = 7.19DD421 pKa = 3.49MFIGGWGTVTMDD433 pKa = 4.33ADD435 pKa = 3.67YY436 pKa = 11.45GLYY439 pKa = 10.09PLFHH443 pKa = 7.18SSGIGNSGNRR453 pKa = 11.84SRR455 pKa = 11.84YY456 pKa = 9.66ANEE459 pKa = 3.89EE460 pKa = 3.78VDD462 pKa = 4.05TLLEE466 pKa = 4.17DD467 pKa = 4.06ARR469 pKa = 11.84TEE471 pKa = 3.74TDD473 pKa = 2.98EE474 pKa = 4.21AQRR477 pKa = 11.84LALYY481 pKa = 10.36EE482 pKa = 4.02EE483 pKa = 4.31AQEE486 pKa = 4.29IIIAEE491 pKa = 4.01APIIPIYY498 pKa = 10.39HH499 pKa = 7.35PYY501 pKa = 10.79LLTGMSEE508 pKa = 4.95DD509 pKa = 3.17IDD511 pKa = 5.17GYY513 pKa = 9.97AQHH516 pKa = 7.11PASFHH521 pKa = 5.79FLSDD525 pKa = 3.43VSKK528 pKa = 10.69EE529 pKa = 3.94
MM1 pKa = 7.6KK2 pKa = 10.2KK3 pKa = 10.49YY4 pKa = 10.75LILVLSLVFVLAACTDD20 pKa = 3.74DD21 pKa = 5.31GEE23 pKa = 4.36VDD25 pKa = 3.25EE26 pKa = 4.88ATEE29 pKa = 4.1EE30 pKa = 4.21EE31 pKa = 4.64ATGEE35 pKa = 4.04DD36 pKa = 3.67AAEE39 pKa = 4.36EE40 pKa = 4.8GSTGGSEE47 pKa = 4.14GTINMSLLATPNSLDD62 pKa = 3.18PHH64 pKa = 6.75AANDD68 pKa = 3.63QPSNHH73 pKa = 5.51VNVNIYY79 pKa = 9.9EE80 pKa = 4.19RR81 pKa = 11.84LVEE84 pKa = 4.3FTPEE88 pKa = 3.95LEE90 pKa = 4.45LEE92 pKa = 4.58PGLAEE97 pKa = 4.44SYY99 pKa = 10.5EE100 pKa = 4.15QVEE103 pKa = 4.35DD104 pKa = 3.98TTWEE108 pKa = 3.8FSIRR112 pKa = 11.84EE113 pKa = 4.16GVSFHH118 pKa = 7.68DD119 pKa = 4.24GEE121 pKa = 4.34EE122 pKa = 4.21LNAEE126 pKa = 4.0AVKK129 pKa = 11.03ANLDD133 pKa = 3.53RR134 pKa = 11.84VTDD137 pKa = 3.83EE138 pKa = 5.32DD139 pKa = 3.79VGSPVAFLFEE149 pKa = 5.19LIEE152 pKa = 4.12EE153 pKa = 4.45VEE155 pKa = 4.51VIDD158 pKa = 5.63DD159 pKa = 4.04YY160 pKa = 11.32TVHH163 pKa = 6.38IHH165 pKa = 5.78TSSPFAALPSHH176 pKa = 6.99LAHH179 pKa = 7.24PAGGMISPAVIEE191 pKa = 3.89EE192 pKa = 4.75DD193 pKa = 3.84YY194 pKa = 11.8AEE196 pKa = 5.04DD197 pKa = 3.71EE198 pKa = 4.34QLTAVHH204 pKa = 5.94QNPVGTGAYY213 pKa = 8.83QFEE216 pKa = 4.91EE217 pKa = 4.42ISEE220 pKa = 4.18GDD222 pKa = 3.73YY223 pKa = 10.0VTLVKK228 pKa = 10.87NEE230 pKa = 4.25EE231 pKa = 4.22YY232 pKa = 9.71WGEE235 pKa = 3.93EE236 pKa = 3.95AEE238 pKa = 4.29SDD240 pKa = 3.85VFTFNAVPEE249 pKa = 4.44DD250 pKa = 3.42ATRR253 pKa = 11.84IAEE256 pKa = 4.17LQTRR260 pKa = 11.84HH261 pKa = 6.38ADD263 pKa = 3.99LIFPLDD269 pKa = 4.02PNDD272 pKa = 3.78FEE274 pKa = 5.63QINSDD279 pKa = 2.94EE280 pKa = 4.17GTTVNEE286 pKa = 4.3TEE288 pKa = 4.51SVRR291 pKa = 11.84MEE293 pKa = 3.64YY294 pKa = 10.69VGFNVEE300 pKa = 4.59VEE302 pKa = 4.31PFDD305 pKa = 5.19DD306 pKa = 3.96PVVRR310 pKa = 11.84QAIAHH315 pKa = 6.97AIDD318 pKa = 4.37KK319 pKa = 10.62EE320 pKa = 4.32DD321 pKa = 4.4IINIMLEE328 pKa = 4.07GKK330 pKa = 10.29AVVADD335 pKa = 4.11TLLSPAVFGHH345 pKa = 6.97AEE347 pKa = 4.49DD348 pKa = 4.19IDD350 pKa = 4.83GIGYY354 pKa = 9.75DD355 pKa = 4.23LEE357 pKa = 4.56TAQEE361 pKa = 4.07LLDD364 pKa = 4.31DD365 pKa = 4.31NGYY368 pKa = 10.76EE369 pKa = 4.06EE370 pKa = 5.3GFSAEE375 pKa = 4.02IVVQDD380 pKa = 3.88RR381 pKa = 11.84TAADD385 pKa = 2.99IATYY389 pKa = 9.92IQEE392 pKa = 4.21QLQEE396 pKa = 4.19LNIDD400 pKa = 3.72LEE402 pKa = 4.74IYY404 pKa = 10.56QIDD407 pKa = 3.51PGAYY411 pKa = 8.92LDD413 pKa = 4.05YY414 pKa = 10.83VGSGSHH420 pKa = 7.19DD421 pKa = 3.49MFIGGWGTVTMDD433 pKa = 4.33ADD435 pKa = 3.67YY436 pKa = 11.45GLYY439 pKa = 10.09PLFHH443 pKa = 7.18SSGIGNSGNRR453 pKa = 11.84SRR455 pKa = 11.84YY456 pKa = 9.66ANEE459 pKa = 3.89EE460 pKa = 3.78VDD462 pKa = 4.05TLLEE466 pKa = 4.17DD467 pKa = 4.06ARR469 pKa = 11.84TEE471 pKa = 3.74TDD473 pKa = 2.98EE474 pKa = 4.21AQRR477 pKa = 11.84LALYY481 pKa = 10.36EE482 pKa = 4.02EE483 pKa = 4.31AQEE486 pKa = 4.29IIIAEE491 pKa = 4.01APIIPIYY498 pKa = 10.39HH499 pKa = 7.35PYY501 pKa = 10.79LLTGMSEE508 pKa = 4.95DD509 pKa = 3.17IDD511 pKa = 5.17GYY513 pKa = 9.97AQHH516 pKa = 7.11PASFHH521 pKa = 5.79FLSDD525 pKa = 3.43VSKK528 pKa = 10.69EE529 pKa = 3.94
Molecular weight: 58.3 kDa
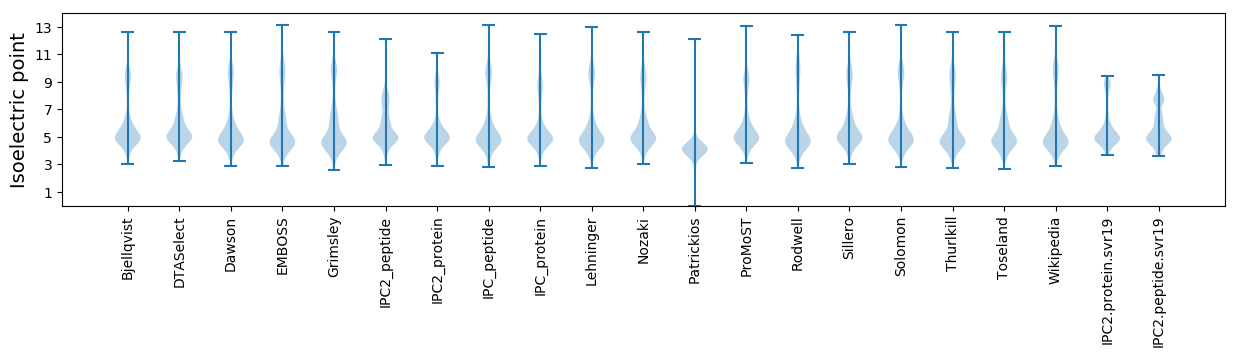
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M7GJP9|A0A1M7GJP9_9STAP Uncharacterized protein OS=Salinicoccus alkaliphilus DSM 16010 OX=1123231 GN=SAMN02745189_01663 PE=4 SV=1
MM1 pKa = 7.62TKK3 pKa = 9.09RR4 pKa = 11.84TYY6 pKa = 10.29QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.87VHH17 pKa = 5.77GFRR20 pKa = 11.84KK21 pKa = 10.02RR22 pKa = 11.84MSTKK26 pKa = 10.06NGRR29 pKa = 11.84KK30 pKa = 8.47ILARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.09GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.62TKK3 pKa = 9.09RR4 pKa = 11.84TYY6 pKa = 10.29QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.87VHH17 pKa = 5.77GFRR20 pKa = 11.84KK21 pKa = 10.02RR22 pKa = 11.84MSTKK26 pKa = 10.06NGRR29 pKa = 11.84KK30 pKa = 8.47ILARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.09GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
753143 |
40 |
1488 |
299.8 |
33.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.3 ± 0.052 | 0.523 ± 0.011 |
6.241 ± 0.051 | 8.244 ± 0.073 |
4.461 ± 0.044 | 7.406 ± 0.05 |
2.313 ± 0.028 | 7.688 ± 0.053 |
5.739 ± 0.051 | 9.258 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.134 ± 0.024 | 4.139 ± 0.033 |
3.502 ± 0.026 | 2.851 ± 0.027 |
4.426 ± 0.042 | 5.762 ± 0.03 |
5.44 ± 0.027 | 7.177 ± 0.036 |
0.804 ± 0.015 | 3.592 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
