
Acidobacterium capsulatum (strain ATCC 51196 / DSM 11244 / JCM 7670 / NBRC 15755 / NCIMB 13165 / 161)
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Acidobacterium; Acidobacterium capsulatum
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
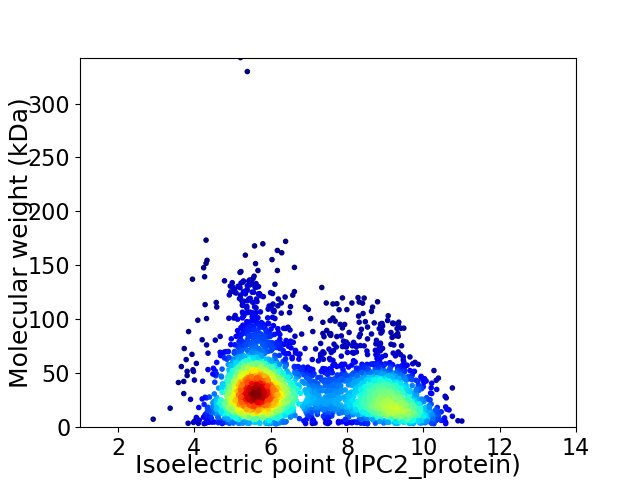
Virtual 2D-PAGE plot for 3363 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C1F943|C1F943_ACIC5 Beta-glucosidase OS=Acidobacterium capsulatum (strain ATCC 51196 / DSM 11244 / JCM 7670 / NBRC 15755 / NCIMB 13165 / 161) OX=240015 GN=ACP_2108 PE=3 SV=1
MM1 pKa = 7.23LTCSQDD7 pKa = 3.46SPSSLLSFFKK17 pKa = 10.77ARR19 pKa = 11.84SAPSRR24 pKa = 11.84LLLVALCSLGLVLAGCSGGSSSSGSSSTSGTAPGGGSGSGGGSNNTPTPGAANSVQLGTATVFDD88 pKa = 4.67GPNPEE93 pKa = 4.27PAQIAADD100 pKa = 3.61ASGDD104 pKa = 3.79DD105 pKa = 3.69TLAWTDD111 pKa = 3.25PSGQVYY117 pKa = 9.47VSRR120 pKa = 11.84LAAGSTSWTTPVALIPATSTLKK142 pKa = 10.52AHH144 pKa = 6.89GGITFAASDD153 pKa = 3.51ASGNTTVLFQASDD166 pKa = 3.42ASFQSYY172 pKa = 9.47LVASTLLAGSSKK184 pKa = 9.7WSQPATLSIPDD195 pKa = 3.83ASGSAATLMSNGDD208 pKa = 2.8IAAAYY213 pKa = 9.37GGSLTDD219 pKa = 3.63SVQVATFAPQTQSWGAPVTAAQSPTLLGNFTITSTPQNALTVLWTANIVPNNDD272 pKa = 3.47PSQQSASGVYY282 pKa = 9.85AATASGSAGPFSTPVEE298 pKa = 4.01IDD300 pKa = 3.49VQPGGTAPGIVTSLPDD316 pKa = 3.53AVTDD320 pKa = 3.65TSGNTTAIWTASPQAGGATDD340 pKa = 5.18LYY342 pKa = 11.03ASRR345 pKa = 11.84LAAGSSTWSKK355 pKa = 10.36PVQLDD360 pKa = 3.59TSSSNNPYY368 pKa = 10.65VGFPTPLVVDD378 pKa = 3.96AQGNVTVAWIAGINSPRR395 pKa = 11.84TVQAVRR401 pKa = 11.84YY402 pKa = 8.12DD403 pKa = 3.34ASTSQWSTPSLVQTLPSGTVAGIPALAIGPTQGEE437 pKa = 4.18VSVFWNQPPSASSSGSNGDD456 pKa = 3.02IYY458 pKa = 11.15QSIWSSGSGSWGNPIQADD476 pKa = 4.21LPLPTAGASAATFPVAAVNGSSSMAVAWGDD506 pKa = 3.5PVTGSNSSYY515 pKa = 10.19TLYY518 pKa = 11.37ANVLQQ523 pKa = 4.54
MM1 pKa = 7.23LTCSQDD7 pKa = 3.46SPSSLLSFFKK17 pKa = 10.77ARR19 pKa = 11.84SAPSRR24 pKa = 11.84LLLVALCSLGLVLAGCSGGSSSSGSSSTSGTAPGGGSGSGGGSNNTPTPGAANSVQLGTATVFDD88 pKa = 4.67GPNPEE93 pKa = 4.27PAQIAADD100 pKa = 3.61ASGDD104 pKa = 3.79DD105 pKa = 3.69TLAWTDD111 pKa = 3.25PSGQVYY117 pKa = 9.47VSRR120 pKa = 11.84LAAGSTSWTTPVALIPATSTLKK142 pKa = 10.52AHH144 pKa = 6.89GGITFAASDD153 pKa = 3.51ASGNTTVLFQASDD166 pKa = 3.42ASFQSYY172 pKa = 9.47LVASTLLAGSSKK184 pKa = 9.7WSQPATLSIPDD195 pKa = 3.83ASGSAATLMSNGDD208 pKa = 2.8IAAAYY213 pKa = 9.37GGSLTDD219 pKa = 3.63SVQVATFAPQTQSWGAPVTAAQSPTLLGNFTITSTPQNALTVLWTANIVPNNDD272 pKa = 3.47PSQQSASGVYY282 pKa = 9.85AATASGSAGPFSTPVEE298 pKa = 4.01IDD300 pKa = 3.49VQPGGTAPGIVTSLPDD316 pKa = 3.53AVTDD320 pKa = 3.65TSGNTTAIWTASPQAGGATDD340 pKa = 5.18LYY342 pKa = 11.03ASRR345 pKa = 11.84LAAGSSTWSKK355 pKa = 10.36PVQLDD360 pKa = 3.59TSSSNNPYY368 pKa = 10.65VGFPTPLVVDD378 pKa = 3.96AQGNVTVAWIAGINSPRR395 pKa = 11.84TVQAVRR401 pKa = 11.84YY402 pKa = 8.12DD403 pKa = 3.34ASTSQWSTPSLVQTLPSGTVAGIPALAIGPTQGEE437 pKa = 4.18VSVFWNQPPSASSSGSNGDD456 pKa = 3.02IYY458 pKa = 11.15QSIWSSGSGSWGNPIQADD476 pKa = 4.21LPLPTAGASAATFPVAAVNGSSSMAVAWGDD506 pKa = 3.5PVTGSNSSYY515 pKa = 10.19TLYY518 pKa = 11.37ANVLQQ523 pKa = 4.54
Molecular weight: 51.78 kDa
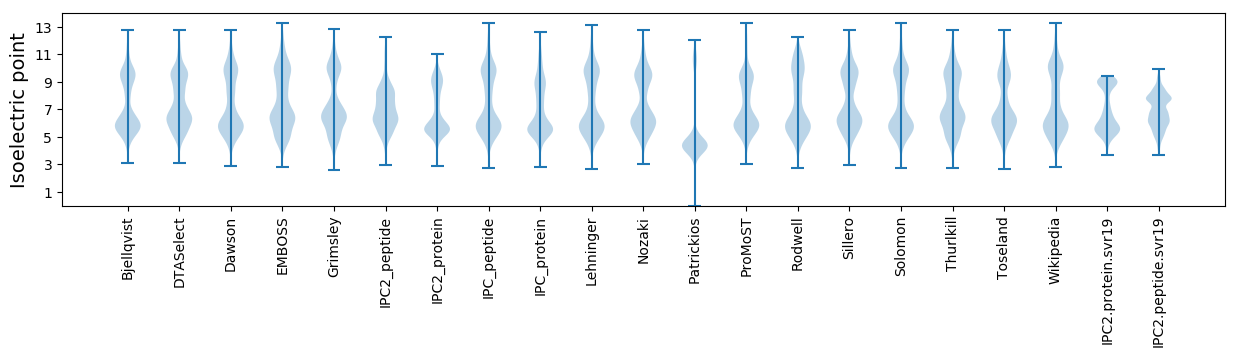
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C1F1H2|C1F1H2_ACIC5 Glucose-6-phosphate 1-dehydrogenase OS=Acidobacterium capsulatum (strain ATCC 51196 / DSM 11244 / JCM 7670 / NBRC 15755 / NCIMB 13165 / 161) OX=240015 GN=zwf1 PE=3 SV=1
MM1 pKa = 7.7ALTRR5 pKa = 11.84HH6 pKa = 5.36ILLRR10 pKa = 11.84NGAFALVGTAVIPAFLTRR28 pKa = 11.84TLHH31 pKa = 5.83AQTAAARR38 pKa = 11.84AWQGAGGGVSARR50 pKa = 11.84RR51 pKa = 11.84GPSRR55 pKa = 3.71
MM1 pKa = 7.7ALTRR5 pKa = 11.84HH6 pKa = 5.36ILLRR10 pKa = 11.84NGAFALVGTAVIPAFLTRR28 pKa = 11.84TLHH31 pKa = 5.83AQTAAARR38 pKa = 11.84AWQGAGGGVSARR50 pKa = 11.84RR51 pKa = 11.84GPSRR55 pKa = 3.71
Molecular weight: 5.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1183035 |
30 |
3102 |
351.8 |
38.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.3 ± 0.054 | 0.905 ± 0.013 |
4.752 ± 0.027 | 5.591 ± 0.047 |
3.775 ± 0.029 | 7.904 ± 0.043 |
2.588 ± 0.021 | 4.855 ± 0.029 |
3.389 ± 0.032 | 10.18 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.37 ± 0.021 | 3.222 ± 0.04 |
5.439 ± 0.034 | 4.25 ± 0.033 |
6.533 ± 0.044 | 6.09 ± 0.044 |
5.62 ± 0.041 | 7.019 ± 0.032 |
1.406 ± 0.019 | 2.811 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
