
Drancourtella sp. An57
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Drancourtella; unclassified Drancourtella
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
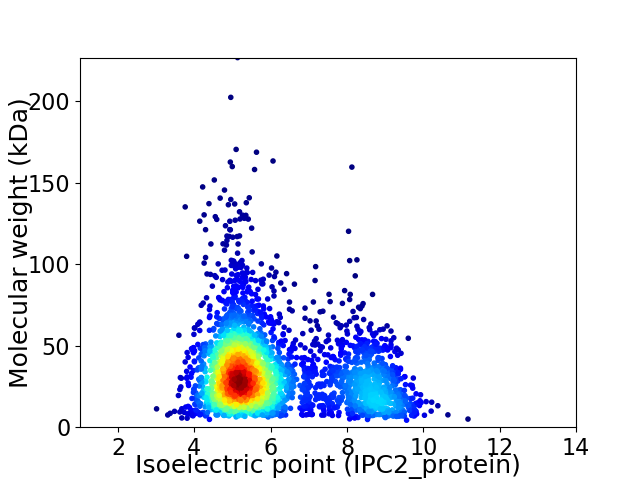
Virtual 2D-PAGE plot for 3069 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y3WA39|A0A1Y3WA39_9FIRM 50S ribosomal protein L2 OS=Drancourtella sp. An57 OX=1965647 GN=rplB PE=3 SV=1
MM1 pKa = 7.18KK2 pKa = 10.33RR3 pKa = 11.84KK4 pKa = 9.33LAVVCIMMLSATTILSGCGGSSDD27 pKa = 4.61DD28 pKa = 4.12SSSGGKK34 pKa = 7.9TKK36 pKa = 10.25IRR38 pKa = 11.84FATWDD43 pKa = 3.34VAEE46 pKa = 5.53DD47 pKa = 3.33VDD49 pKa = 4.44KK50 pKa = 11.31QQEE53 pKa = 4.19LVDD56 pKa = 4.19KK57 pKa = 10.82FNEE60 pKa = 3.91EE61 pKa = 3.76HH62 pKa = 7.68DD63 pKa = 4.91DD64 pKa = 3.99IEE66 pKa = 4.66VTLEE70 pKa = 4.04AYY72 pKa = 10.28GSDD75 pKa = 3.71FDD77 pKa = 4.38TKK79 pKa = 10.34ISAGMGSGDD88 pKa = 4.1TPDD91 pKa = 4.25VMYY94 pKa = 9.35MWNYY98 pKa = 7.71PAYY101 pKa = 10.07YY102 pKa = 10.18DD103 pKa = 4.15GLEE106 pKa = 4.33PLDD109 pKa = 5.09DD110 pKa = 4.73YY111 pKa = 11.25IAEE114 pKa = 4.57EE115 pKa = 4.14GDD117 pKa = 3.64EE118 pKa = 4.32YY119 pKa = 11.44KK120 pKa = 11.26SNFYY124 pKa = 8.72NTLWDD129 pKa = 3.96YY130 pKa = 12.15NMMDD134 pKa = 3.36GSTYY138 pKa = 10.28GIPVGFTTHH147 pKa = 6.58CLFYY151 pKa = 11.12NKK153 pKa = 10.23DD154 pKa = 3.3IFDD157 pKa = 4.11EE158 pKa = 4.85AGVEE162 pKa = 4.18YY163 pKa = 8.47PTADD167 pKa = 3.53WTWDD171 pKa = 3.66DD172 pKa = 3.95LQAAAKK178 pKa = 8.7TISEE182 pKa = 4.22KK183 pKa = 10.28TDD185 pKa = 2.87AKK187 pKa = 10.95GFSFQMKK194 pKa = 8.93PDD196 pKa = 4.18PYY198 pKa = 10.78DD199 pKa = 3.45FEE201 pKa = 4.59MYY203 pKa = 10.03LWSNGTSYY211 pKa = 11.19CDD213 pKa = 3.59EE214 pKa = 4.41NGEE217 pKa = 3.78MDD219 pKa = 4.36GYY221 pKa = 10.6INSEE225 pKa = 3.79EE226 pKa = 4.17SQEE229 pKa = 4.0VFQMFQDD236 pKa = 3.93MEE238 pKa = 4.4EE239 pKa = 4.1EE240 pKa = 5.31GYY242 pKa = 8.77ATATEE247 pKa = 4.33GDD249 pKa = 3.66GTDD252 pKa = 3.34EE253 pKa = 4.09FRR255 pKa = 11.84AGSTAMYY262 pKa = 10.37VYY264 pKa = 10.57GSWSINTLNEE274 pKa = 4.14DD275 pKa = 3.49GVNYY279 pKa = 10.41GVTTIPAFADD289 pKa = 3.34AGQDD293 pKa = 3.47SVSILSSSGLSMSKK307 pKa = 10.57DD308 pKa = 3.69SEE310 pKa = 4.45HH311 pKa = 7.4KK312 pKa = 9.93DD313 pKa = 3.39AAWEE317 pKa = 4.51FIKK320 pKa = 11.0YY321 pKa = 6.25WTNEE325 pKa = 3.74EE326 pKa = 4.18CNKK329 pKa = 10.15ARR331 pKa = 11.84IGTEE335 pKa = 3.9LPVLNTVVEE344 pKa = 4.5SEE346 pKa = 4.59GIMEE350 pKa = 4.18QEE352 pKa = 4.43EE353 pKa = 4.43YY354 pKa = 11.38APFYY358 pKa = 11.15TMLEE362 pKa = 4.15QSSGHH367 pKa = 5.65TPASFLMEE375 pKa = 4.12SWSEE379 pKa = 3.9VSEE382 pKa = 4.01NLSLSFEE389 pKa = 4.47QIFNPSSMQDD399 pKa = 3.02VSEE402 pKa = 4.4VLDD405 pKa = 3.87SAVKK409 pKa = 10.31
MM1 pKa = 7.18KK2 pKa = 10.33RR3 pKa = 11.84KK4 pKa = 9.33LAVVCIMMLSATTILSGCGGSSDD27 pKa = 4.61DD28 pKa = 4.12SSSGGKK34 pKa = 7.9TKK36 pKa = 10.25IRR38 pKa = 11.84FATWDD43 pKa = 3.34VAEE46 pKa = 5.53DD47 pKa = 3.33VDD49 pKa = 4.44KK50 pKa = 11.31QQEE53 pKa = 4.19LVDD56 pKa = 4.19KK57 pKa = 10.82FNEE60 pKa = 3.91EE61 pKa = 3.76HH62 pKa = 7.68DD63 pKa = 4.91DD64 pKa = 3.99IEE66 pKa = 4.66VTLEE70 pKa = 4.04AYY72 pKa = 10.28GSDD75 pKa = 3.71FDD77 pKa = 4.38TKK79 pKa = 10.34ISAGMGSGDD88 pKa = 4.1TPDD91 pKa = 4.25VMYY94 pKa = 9.35MWNYY98 pKa = 7.71PAYY101 pKa = 10.07YY102 pKa = 10.18DD103 pKa = 4.15GLEE106 pKa = 4.33PLDD109 pKa = 5.09DD110 pKa = 4.73YY111 pKa = 11.25IAEE114 pKa = 4.57EE115 pKa = 4.14GDD117 pKa = 3.64EE118 pKa = 4.32YY119 pKa = 11.44KK120 pKa = 11.26SNFYY124 pKa = 8.72NTLWDD129 pKa = 3.96YY130 pKa = 12.15NMMDD134 pKa = 3.36GSTYY138 pKa = 10.28GIPVGFTTHH147 pKa = 6.58CLFYY151 pKa = 11.12NKK153 pKa = 10.23DD154 pKa = 3.3IFDD157 pKa = 4.11EE158 pKa = 4.85AGVEE162 pKa = 4.18YY163 pKa = 8.47PTADD167 pKa = 3.53WTWDD171 pKa = 3.66DD172 pKa = 3.95LQAAAKK178 pKa = 8.7TISEE182 pKa = 4.22KK183 pKa = 10.28TDD185 pKa = 2.87AKK187 pKa = 10.95GFSFQMKK194 pKa = 8.93PDD196 pKa = 4.18PYY198 pKa = 10.78DD199 pKa = 3.45FEE201 pKa = 4.59MYY203 pKa = 10.03LWSNGTSYY211 pKa = 11.19CDD213 pKa = 3.59EE214 pKa = 4.41NGEE217 pKa = 3.78MDD219 pKa = 4.36GYY221 pKa = 10.6INSEE225 pKa = 3.79EE226 pKa = 4.17SQEE229 pKa = 4.0VFQMFQDD236 pKa = 3.93MEE238 pKa = 4.4EE239 pKa = 4.1EE240 pKa = 5.31GYY242 pKa = 8.77ATATEE247 pKa = 4.33GDD249 pKa = 3.66GTDD252 pKa = 3.34EE253 pKa = 4.09FRR255 pKa = 11.84AGSTAMYY262 pKa = 10.37VYY264 pKa = 10.57GSWSINTLNEE274 pKa = 4.14DD275 pKa = 3.49GVNYY279 pKa = 10.41GVTTIPAFADD289 pKa = 3.34AGQDD293 pKa = 3.47SVSILSSSGLSMSKK307 pKa = 10.57DD308 pKa = 3.69SEE310 pKa = 4.45HH311 pKa = 7.4KK312 pKa = 9.93DD313 pKa = 3.39AAWEE317 pKa = 4.51FIKK320 pKa = 11.0YY321 pKa = 6.25WTNEE325 pKa = 3.74EE326 pKa = 4.18CNKK329 pKa = 10.15ARR331 pKa = 11.84IGTEE335 pKa = 3.9LPVLNTVVEE344 pKa = 4.5SEE346 pKa = 4.59GIMEE350 pKa = 4.18QEE352 pKa = 4.43EE353 pKa = 4.43YY354 pKa = 11.38APFYY358 pKa = 11.15TMLEE362 pKa = 4.15QSSGHH367 pKa = 5.65TPASFLMEE375 pKa = 4.12SWSEE379 pKa = 3.9VSEE382 pKa = 4.01NLSLSFEE389 pKa = 4.47QIFNPSSMQDD399 pKa = 3.02VSEE402 pKa = 4.4VLDD405 pKa = 3.87SAVKK409 pKa = 10.31
Molecular weight: 45.79 kDa
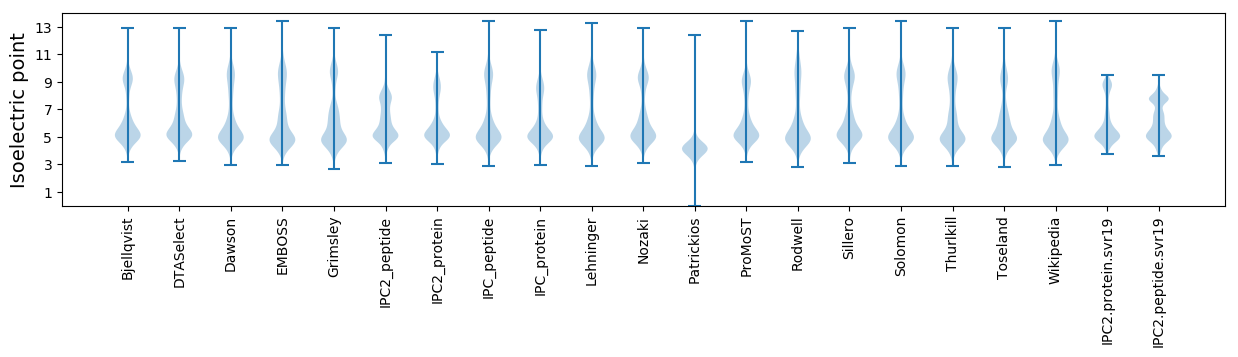
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y3W3X1|A0A1Y3W3X1_9FIRM Alanine--tRNA ligase OS=Drancourtella sp. An57 OX=1965647 GN=B5G11_14770 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 4.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
969582 |
37 |
1974 |
315.9 |
35.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.263 ± 0.047 | 1.47 ± 0.017 |
5.503 ± 0.034 | 8.458 ± 0.061 |
4.064 ± 0.031 | 7.221 ± 0.043 |
1.811 ± 0.02 | 7.142 ± 0.046 |
6.765 ± 0.04 | 8.928 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.141 ± 0.023 | 3.842 ± 0.031 |
3.371 ± 0.026 | 3.376 ± 0.023 |
4.934 ± 0.038 | 5.538 ± 0.039 |
5.316 ± 0.034 | 6.77 ± 0.037 |
0.966 ± 0.015 | 4.119 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
