
Zaire ebolavirus (strain Mayinga-76) (ZEBOV) (Zaire Ebola virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Filoviridae; Ebolavirus; Zaire ebolavirus; Ebola virus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
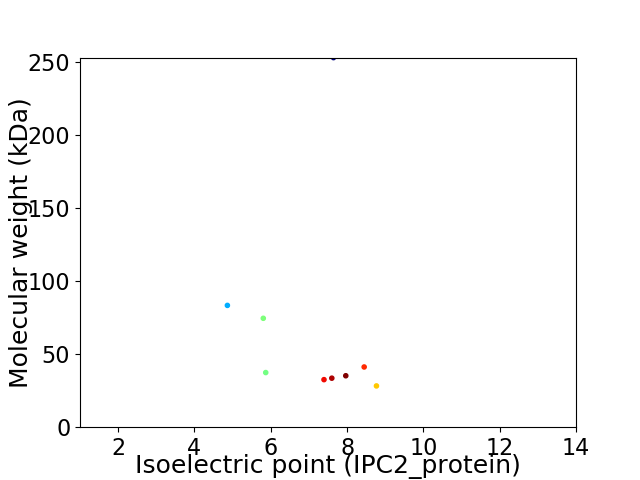
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
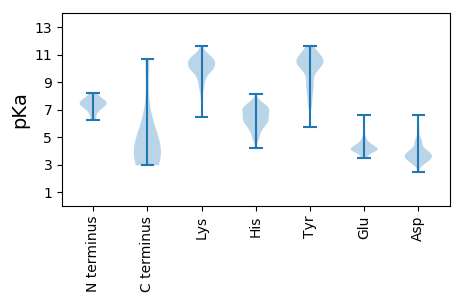
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P60170|VSGP_EBOZM Pre-small/secreted glycoprotein OS=Zaire ebolavirus (strain Mayinga-76) OX=128952 GN=GP PE=1 SV=1
MM1 pKa = 8.24DD2 pKa = 5.39SRR4 pKa = 11.84PQKK7 pKa = 10.28IWMAPSLTEE16 pKa = 4.62SDD18 pKa = 3.12MDD20 pKa = 3.5YY21 pKa = 11.37HH22 pKa = 7.46KK23 pKa = 10.77ILTAGLSVQQGIVRR37 pKa = 11.84QRR39 pKa = 11.84VIPVYY44 pKa = 9.99QVNNLEE50 pKa = 4.85EE51 pKa = 4.26ICQLIIQAFEE61 pKa = 4.21AGVDD65 pKa = 4.26FQEE68 pKa = 4.79SADD71 pKa = 3.83SFLLMLCLHH80 pKa = 7.05HH81 pKa = 7.47AYY83 pKa = 10.35QGDD86 pKa = 3.72YY87 pKa = 11.27KK88 pKa = 11.19LFLEE92 pKa = 4.69SGAVKK97 pKa = 10.45YY98 pKa = 11.09LEE100 pKa = 3.87GHH102 pKa = 5.74GFRR105 pKa = 11.84FEE107 pKa = 4.07VKK109 pKa = 10.23KK110 pKa = 10.7RR111 pKa = 11.84DD112 pKa = 3.34GVKK115 pKa = 10.25RR116 pKa = 11.84LEE118 pKa = 4.21EE119 pKa = 3.99LLPAVSSGKK128 pKa = 10.01NIKK131 pKa = 8.74RR132 pKa = 11.84TLAAMPEE139 pKa = 4.26EE140 pKa = 4.61EE141 pKa = 4.34TTEE144 pKa = 4.02ANAGQFLSFASLFLPKK160 pKa = 10.39LVVGEE165 pKa = 4.31KK166 pKa = 10.46ACLEE170 pKa = 4.0KK171 pKa = 10.81VQRR174 pKa = 11.84QIQVHH179 pKa = 6.09AEE181 pKa = 3.78QGLIQYY187 pKa = 7.32PTAWQSVGHH196 pKa = 5.52MMVIFRR202 pKa = 11.84LMRR205 pKa = 11.84TNFLIKK211 pKa = 10.22FLLIHH216 pKa = 6.24QGMHH220 pKa = 5.09MVAGHH225 pKa = 7.11DD226 pKa = 3.84ANDD229 pKa = 3.63AVISNSVAQARR240 pKa = 11.84FSGLLIVKK248 pKa = 8.23TVLDD252 pKa = 4.78HH253 pKa = 7.38ILQKK257 pKa = 8.28TEE259 pKa = 3.67RR260 pKa = 11.84GVRR263 pKa = 11.84LHH265 pKa = 6.46PLARR269 pKa = 11.84TAKK272 pKa = 9.83VKK274 pKa = 11.22NEE276 pKa = 3.83VNSFKK281 pKa = 10.95AALSSLAKK289 pKa = 9.86HH290 pKa = 6.16GEE292 pKa = 3.99YY293 pKa = 10.9APFARR298 pKa = 11.84LLNLSGVNNLEE309 pKa = 4.48HH310 pKa = 7.06GLFPQLSAIALGVATAHH327 pKa = 6.58GSTLAGVNVGEE338 pKa = 4.64QYY340 pKa = 10.17QQLRR344 pKa = 11.84EE345 pKa = 4.06AATEE349 pKa = 4.1AEE351 pKa = 4.44KK352 pKa = 10.74QLQQYY357 pKa = 10.35AEE359 pKa = 4.16SRR361 pKa = 11.84EE362 pKa = 3.99LDD364 pKa = 3.55HH365 pKa = 7.43LGLDD369 pKa = 3.83DD370 pKa = 4.06QEE372 pKa = 5.56KK373 pKa = 9.82KK374 pKa = 10.65ILMNFHH380 pKa = 5.72QKK382 pKa = 10.38KK383 pKa = 10.33NEE385 pKa = 3.58ISFQQTNAMVTLRR398 pKa = 11.84KK399 pKa = 9.42EE400 pKa = 3.98RR401 pKa = 11.84LAKK404 pKa = 9.27LTEE407 pKa = 5.11AITAASLPKK416 pKa = 10.33TSGHH420 pKa = 6.29YY421 pKa = 10.7DD422 pKa = 3.75DD423 pKa = 6.46DD424 pKa = 6.63DD425 pKa = 6.0DD426 pKa = 5.55IPFPGPINDD435 pKa = 4.78DD436 pKa = 4.41DD437 pKa = 4.81NPGHH441 pKa = 6.58QDD443 pKa = 4.06DD444 pKa = 5.69DD445 pKa = 4.13PTDD448 pKa = 3.87SQDD451 pKa = 3.01TTIPDD456 pKa = 3.71VVVDD460 pKa = 4.75PDD462 pKa = 3.79DD463 pKa = 4.14GSYY466 pKa = 11.56GEE468 pKa = 4.07YY469 pKa = 10.17QSYY472 pKa = 10.58SEE474 pKa = 4.71NGMNAPDD481 pKa = 4.41DD482 pKa = 4.49LVLFDD487 pKa = 6.03LDD489 pKa = 5.11EE490 pKa = 6.61DD491 pKa = 4.98DD492 pKa = 5.69EE493 pKa = 4.56DD494 pKa = 4.23TKK496 pKa = 11.0PVPNRR501 pKa = 11.84STKK504 pKa = 10.49GGQQKK509 pKa = 10.34NSQKK513 pKa = 9.96GQHH516 pKa = 5.72IEE518 pKa = 3.92GRR520 pKa = 11.84QTQSRR525 pKa = 11.84PIQNVPGPHH534 pKa = 5.51RR535 pKa = 11.84TIHH538 pKa = 6.16HH539 pKa = 6.92ASAPLTDD546 pKa = 3.32NDD548 pKa = 4.4RR549 pKa = 11.84RR550 pKa = 11.84NEE552 pKa = 4.06PSGSTSPRR560 pKa = 11.84MLTPINEE567 pKa = 4.36EE568 pKa = 4.16ADD570 pKa = 3.9PLDD573 pKa = 4.54DD574 pKa = 6.08ADD576 pKa = 5.93DD577 pKa = 4.21EE578 pKa = 4.83TSSLPPLEE586 pKa = 5.6SDD588 pKa = 5.56DD589 pKa = 4.71EE590 pKa = 4.3EE591 pKa = 4.48QDD593 pKa = 3.9RR594 pKa = 11.84DD595 pKa = 3.69GTSNRR600 pKa = 11.84TPTVAPPAPVYY611 pKa = 10.21RR612 pKa = 11.84DD613 pKa = 3.24HH614 pKa = 7.49SEE616 pKa = 4.16KK617 pKa = 10.78KK618 pKa = 9.94EE619 pKa = 3.98LPQDD623 pKa = 3.52EE624 pKa = 4.87QQDD627 pKa = 3.52QDD629 pKa = 3.46HH630 pKa = 5.74TQEE633 pKa = 4.96ARR635 pKa = 11.84NQDD638 pKa = 3.18SDD640 pKa = 3.73NTQSEE645 pKa = 4.51HH646 pKa = 7.33SFEE649 pKa = 3.72EE650 pKa = 4.72MYY652 pKa = 10.9RR653 pKa = 11.84HH654 pKa = 6.72ILRR657 pKa = 11.84SQGPFDD663 pKa = 3.59AVLYY667 pKa = 8.01YY668 pKa = 11.21HH669 pKa = 6.23MMKK672 pKa = 10.56DD673 pKa = 3.48EE674 pKa = 4.43PVVFSTSDD682 pKa = 3.15GKK684 pKa = 11.22EE685 pKa = 3.56YY686 pKa = 10.03TYY688 pKa = 10.88PDD690 pKa = 3.91SLEE693 pKa = 4.06EE694 pKa = 4.86EE695 pKa = 4.48YY696 pKa = 10.61PPWLTEE702 pKa = 3.81KK703 pKa = 10.66EE704 pKa = 4.2AMNEE708 pKa = 3.64EE709 pKa = 4.19NRR711 pKa = 11.84FVTLDD716 pKa = 3.26GQQFYY721 pKa = 9.84WPVMNHH727 pKa = 5.74KK728 pKa = 9.96NKK730 pKa = 9.79FMAILQHH737 pKa = 5.92HH738 pKa = 5.25QQ739 pKa = 2.95
MM1 pKa = 8.24DD2 pKa = 5.39SRR4 pKa = 11.84PQKK7 pKa = 10.28IWMAPSLTEE16 pKa = 4.62SDD18 pKa = 3.12MDD20 pKa = 3.5YY21 pKa = 11.37HH22 pKa = 7.46KK23 pKa = 10.77ILTAGLSVQQGIVRR37 pKa = 11.84QRR39 pKa = 11.84VIPVYY44 pKa = 9.99QVNNLEE50 pKa = 4.85EE51 pKa = 4.26ICQLIIQAFEE61 pKa = 4.21AGVDD65 pKa = 4.26FQEE68 pKa = 4.79SADD71 pKa = 3.83SFLLMLCLHH80 pKa = 7.05HH81 pKa = 7.47AYY83 pKa = 10.35QGDD86 pKa = 3.72YY87 pKa = 11.27KK88 pKa = 11.19LFLEE92 pKa = 4.69SGAVKK97 pKa = 10.45YY98 pKa = 11.09LEE100 pKa = 3.87GHH102 pKa = 5.74GFRR105 pKa = 11.84FEE107 pKa = 4.07VKK109 pKa = 10.23KK110 pKa = 10.7RR111 pKa = 11.84DD112 pKa = 3.34GVKK115 pKa = 10.25RR116 pKa = 11.84LEE118 pKa = 4.21EE119 pKa = 3.99LLPAVSSGKK128 pKa = 10.01NIKK131 pKa = 8.74RR132 pKa = 11.84TLAAMPEE139 pKa = 4.26EE140 pKa = 4.61EE141 pKa = 4.34TTEE144 pKa = 4.02ANAGQFLSFASLFLPKK160 pKa = 10.39LVVGEE165 pKa = 4.31KK166 pKa = 10.46ACLEE170 pKa = 4.0KK171 pKa = 10.81VQRR174 pKa = 11.84QIQVHH179 pKa = 6.09AEE181 pKa = 3.78QGLIQYY187 pKa = 7.32PTAWQSVGHH196 pKa = 5.52MMVIFRR202 pKa = 11.84LMRR205 pKa = 11.84TNFLIKK211 pKa = 10.22FLLIHH216 pKa = 6.24QGMHH220 pKa = 5.09MVAGHH225 pKa = 7.11DD226 pKa = 3.84ANDD229 pKa = 3.63AVISNSVAQARR240 pKa = 11.84FSGLLIVKK248 pKa = 8.23TVLDD252 pKa = 4.78HH253 pKa = 7.38ILQKK257 pKa = 8.28TEE259 pKa = 3.67RR260 pKa = 11.84GVRR263 pKa = 11.84LHH265 pKa = 6.46PLARR269 pKa = 11.84TAKK272 pKa = 9.83VKK274 pKa = 11.22NEE276 pKa = 3.83VNSFKK281 pKa = 10.95AALSSLAKK289 pKa = 9.86HH290 pKa = 6.16GEE292 pKa = 3.99YY293 pKa = 10.9APFARR298 pKa = 11.84LLNLSGVNNLEE309 pKa = 4.48HH310 pKa = 7.06GLFPQLSAIALGVATAHH327 pKa = 6.58GSTLAGVNVGEE338 pKa = 4.64QYY340 pKa = 10.17QQLRR344 pKa = 11.84EE345 pKa = 4.06AATEE349 pKa = 4.1AEE351 pKa = 4.44KK352 pKa = 10.74QLQQYY357 pKa = 10.35AEE359 pKa = 4.16SRR361 pKa = 11.84EE362 pKa = 3.99LDD364 pKa = 3.55HH365 pKa = 7.43LGLDD369 pKa = 3.83DD370 pKa = 4.06QEE372 pKa = 5.56KK373 pKa = 9.82KK374 pKa = 10.65ILMNFHH380 pKa = 5.72QKK382 pKa = 10.38KK383 pKa = 10.33NEE385 pKa = 3.58ISFQQTNAMVTLRR398 pKa = 11.84KK399 pKa = 9.42EE400 pKa = 3.98RR401 pKa = 11.84LAKK404 pKa = 9.27LTEE407 pKa = 5.11AITAASLPKK416 pKa = 10.33TSGHH420 pKa = 6.29YY421 pKa = 10.7DD422 pKa = 3.75DD423 pKa = 6.46DD424 pKa = 6.63DD425 pKa = 6.0DD426 pKa = 5.55IPFPGPINDD435 pKa = 4.78DD436 pKa = 4.41DD437 pKa = 4.81NPGHH441 pKa = 6.58QDD443 pKa = 4.06DD444 pKa = 5.69DD445 pKa = 4.13PTDD448 pKa = 3.87SQDD451 pKa = 3.01TTIPDD456 pKa = 3.71VVVDD460 pKa = 4.75PDD462 pKa = 3.79DD463 pKa = 4.14GSYY466 pKa = 11.56GEE468 pKa = 4.07YY469 pKa = 10.17QSYY472 pKa = 10.58SEE474 pKa = 4.71NGMNAPDD481 pKa = 4.41DD482 pKa = 4.49LVLFDD487 pKa = 6.03LDD489 pKa = 5.11EE490 pKa = 6.61DD491 pKa = 4.98DD492 pKa = 5.69EE493 pKa = 4.56DD494 pKa = 4.23TKK496 pKa = 11.0PVPNRR501 pKa = 11.84STKK504 pKa = 10.49GGQQKK509 pKa = 10.34NSQKK513 pKa = 9.96GQHH516 pKa = 5.72IEE518 pKa = 3.92GRR520 pKa = 11.84QTQSRR525 pKa = 11.84PIQNVPGPHH534 pKa = 5.51RR535 pKa = 11.84TIHH538 pKa = 6.16HH539 pKa = 6.92ASAPLTDD546 pKa = 3.32NDD548 pKa = 4.4RR549 pKa = 11.84RR550 pKa = 11.84NEE552 pKa = 4.06PSGSTSPRR560 pKa = 11.84MLTPINEE567 pKa = 4.36EE568 pKa = 4.16ADD570 pKa = 3.9PLDD573 pKa = 4.54DD574 pKa = 6.08ADD576 pKa = 5.93DD577 pKa = 4.21EE578 pKa = 4.83TSSLPPLEE586 pKa = 5.6SDD588 pKa = 5.56DD589 pKa = 4.71EE590 pKa = 4.3EE591 pKa = 4.48QDD593 pKa = 3.9RR594 pKa = 11.84DD595 pKa = 3.69GTSNRR600 pKa = 11.84TPTVAPPAPVYY611 pKa = 10.21RR612 pKa = 11.84DD613 pKa = 3.24HH614 pKa = 7.49SEE616 pKa = 4.16KK617 pKa = 10.78KK618 pKa = 9.94EE619 pKa = 3.98LPQDD623 pKa = 3.52EE624 pKa = 4.87QQDD627 pKa = 3.52QDD629 pKa = 3.46HH630 pKa = 5.74TQEE633 pKa = 4.96ARR635 pKa = 11.84NQDD638 pKa = 3.18SDD640 pKa = 3.73NTQSEE645 pKa = 4.51HH646 pKa = 7.33SFEE649 pKa = 3.72EE650 pKa = 4.72MYY652 pKa = 10.9RR653 pKa = 11.84HH654 pKa = 6.72ILRR657 pKa = 11.84SQGPFDD663 pKa = 3.59AVLYY667 pKa = 8.01YY668 pKa = 11.21HH669 pKa = 6.23MMKK672 pKa = 10.56DD673 pKa = 3.48EE674 pKa = 4.43PVVFSTSDD682 pKa = 3.15GKK684 pKa = 11.22EE685 pKa = 3.56YY686 pKa = 10.03TYY688 pKa = 10.88PDD690 pKa = 3.91SLEE693 pKa = 4.06EE694 pKa = 4.86EE695 pKa = 4.48YY696 pKa = 10.61PPWLTEE702 pKa = 3.81KK703 pKa = 10.66EE704 pKa = 4.2AMNEE708 pKa = 3.64EE709 pKa = 4.19NRR711 pKa = 11.84FVTLDD716 pKa = 3.26GQQFYY721 pKa = 9.84WPVMNHH727 pKa = 5.74KK728 pKa = 9.96NKK730 pKa = 9.79FMAILQHH737 pKa = 5.92HH738 pKa = 5.25QQ739 pKa = 2.95
Molecular weight: 83.29 kDa
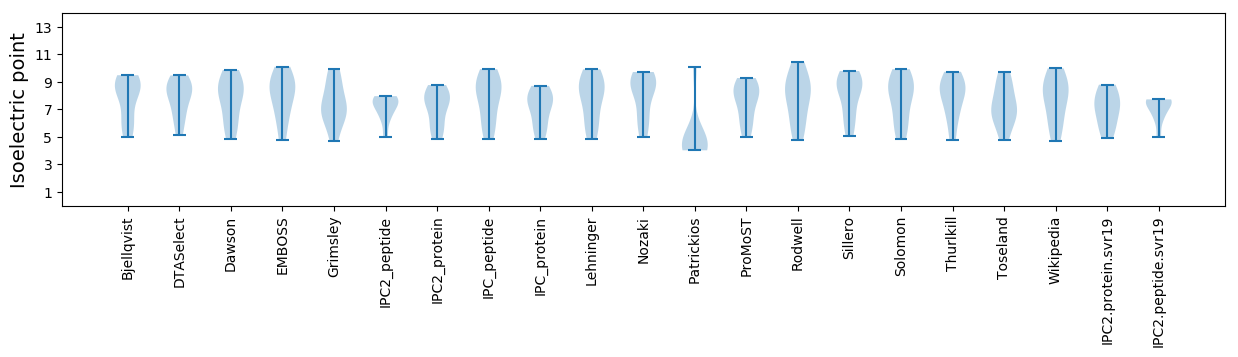
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q05323|VP30_EBOZM Transcriptional activator VP30 OS=Zaire ebolavirus (strain Mayinga-76) OX=128952 GN=VP30 PE=1 SV=2
MM1 pKa = 7.54AKK3 pKa = 9.15ATGRR7 pKa = 11.84YY8 pKa = 9.15NLISPKK14 pKa = 9.97KK15 pKa = 8.31DD16 pKa = 3.15LEE18 pKa = 4.48KK19 pKa = 11.0GVVLSDD25 pKa = 3.51LCNFLVSQTIQGWKK39 pKa = 9.83VYY41 pKa = 8.73WAGIEE46 pKa = 3.85FDD48 pKa = 3.84VTHH51 pKa = 6.93KK52 pKa = 10.92GMALLHH58 pKa = 5.94RR59 pKa = 11.84LKK61 pKa = 10.38TNDD64 pKa = 3.85FAPAWSMTRR73 pKa = 11.84NLFPHH78 pKa = 7.3LFQNPNSTIEE88 pKa = 4.11SPLWALRR95 pKa = 11.84VILAAGIQDD104 pKa = 3.69QLIDD108 pKa = 3.49QSLIEE113 pKa = 4.47PLAGALGLISDD124 pKa = 4.69WLLTTNTNHH133 pKa = 6.45FNMRR137 pKa = 11.84TQRR140 pKa = 11.84VKK142 pKa = 10.77EE143 pKa = 3.89QLSLKK148 pKa = 8.85MLSLIRR154 pKa = 11.84SNILKK159 pKa = 10.16FINKK163 pKa = 9.24LDD165 pKa = 3.88ALHH168 pKa = 5.76VVNYY172 pKa = 10.4NGLLSSIEE180 pKa = 4.19IGTQNHH186 pKa = 6.3TIIITRR192 pKa = 11.84TNMGFLVEE200 pKa = 4.29LQEE203 pKa = 4.39PDD205 pKa = 3.16KK206 pKa = 11.53SAMNRR211 pKa = 11.84MKK213 pKa = 10.51PGPAKK218 pKa = 10.49FSLLHH223 pKa = 6.32EE224 pKa = 4.63STLKK228 pKa = 11.0AFTQGSSTRR237 pKa = 11.84MQSLILEE244 pKa = 4.63FNSSLAII251 pKa = 4.01
MM1 pKa = 7.54AKK3 pKa = 9.15ATGRR7 pKa = 11.84YY8 pKa = 9.15NLISPKK14 pKa = 9.97KK15 pKa = 8.31DD16 pKa = 3.15LEE18 pKa = 4.48KK19 pKa = 11.0GVVLSDD25 pKa = 3.51LCNFLVSQTIQGWKK39 pKa = 9.83VYY41 pKa = 8.73WAGIEE46 pKa = 3.85FDD48 pKa = 3.84VTHH51 pKa = 6.93KK52 pKa = 10.92GMALLHH58 pKa = 5.94RR59 pKa = 11.84LKK61 pKa = 10.38TNDD64 pKa = 3.85FAPAWSMTRR73 pKa = 11.84NLFPHH78 pKa = 7.3LFQNPNSTIEE88 pKa = 4.11SPLWALRR95 pKa = 11.84VILAAGIQDD104 pKa = 3.69QLIDD108 pKa = 3.49QSLIEE113 pKa = 4.47PLAGALGLISDD124 pKa = 4.69WLLTTNTNHH133 pKa = 6.45FNMRR137 pKa = 11.84TQRR140 pKa = 11.84VKK142 pKa = 10.77EE143 pKa = 3.89QLSLKK148 pKa = 8.85MLSLIRR154 pKa = 11.84SNILKK159 pKa = 10.16FINKK163 pKa = 9.24LDD165 pKa = 3.88ALHH168 pKa = 5.76VVNYY172 pKa = 10.4NGLLSSIEE180 pKa = 4.19IGTQNHH186 pKa = 6.3TIIITRR192 pKa = 11.84TNMGFLVEE200 pKa = 4.29LQEE203 pKa = 4.39PDD205 pKa = 3.16KK206 pKa = 11.53SAMNRR211 pKa = 11.84MKK213 pKa = 10.51PGPAKK218 pKa = 10.49FSLLHH223 pKa = 6.32EE224 pKa = 4.63STLKK228 pKa = 11.0AFTQGSSTRR237 pKa = 11.84MQSLILEE244 pKa = 4.63FNSSLAII251 pKa = 4.01
Molecular weight: 28.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5494 |
251 |
2212 |
610.4 |
68.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.17 ± 0.375 | 1.638 ± 0.23 |
5.133 ± 0.484 | 5.533 ± 0.5 |
4.587 ± 0.397 | 5.679 ± 0.572 |
2.967 ± 0.37 | 5.952 ± 0.504 |
5.224 ± 0.201 | 10.084 ± 0.637 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.675 ± 0.276 | 4.605 ± 0.266 |
5.479 ± 0.625 | 5.042 ± 0.454 |
5.206 ± 0.29 | 7.663 ± 0.323 |
7.608 ± 0.644 | 5.388 ± 0.286 |
1.365 ± 0.2 | 3.003 ± 0.441 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
