
Pseudosporangium ferrugineum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Pseudosporangium
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
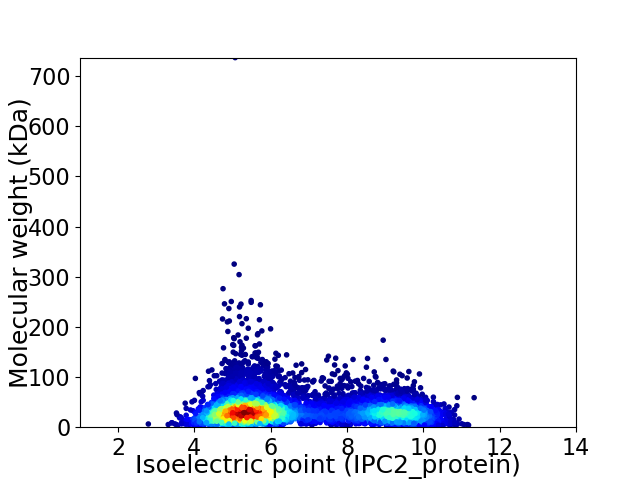
Virtual 2D-PAGE plot for 7729 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0RI42|A0A2T0RI42_9ACTN Dye decolorizing peroxidase OS=Pseudosporangium ferrugineum OX=439699 GN=CLV70_12236 PE=4 SV=1
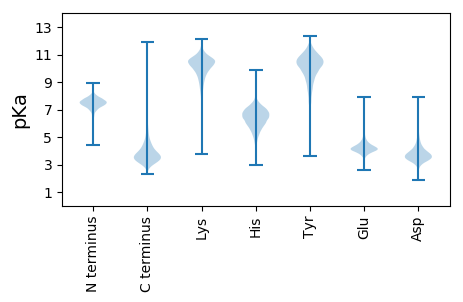
MM1 pKa = 8.03DD2 pKa = 5.63SSPTLQDD9 pKa = 3.27FVLNLIYY16 pKa = 10.8DD17 pKa = 4.28PAARR21 pKa = 11.84SAFEE25 pKa = 4.53LDD27 pKa = 3.62PEE29 pKa = 4.87GVLADD34 pKa = 4.38AGLSDD39 pKa = 3.78VTAVDD44 pKa = 3.73VQEE47 pKa = 4.5VIPLVVDD54 pKa = 3.42YY55 pKa = 11.36APLSDD60 pKa = 3.85VAAVTAPLGVHH71 pKa = 7.1DD72 pKa = 5.36LTTGVADD79 pKa = 4.28ADD81 pKa = 3.7VSAAVAQLQAMTGNLTPGAHH101 pKa = 7.02AAPDD105 pKa = 3.48VAYY108 pKa = 9.95AGAGAFMVSTDD119 pKa = 3.51GLLAGTPAVGVGLEE133 pKa = 4.06HH134 pKa = 7.27AATVDD139 pKa = 3.71APVSTSVTAPVDD151 pKa = 3.64GALSAVHH158 pKa = 7.15DD159 pKa = 4.88PGLGLDD165 pKa = 4.2AGVSTATSVTTTVVGDD181 pKa = 3.86ADD183 pKa = 3.89GLIASTGVDD192 pKa = 3.27AGATLDD198 pKa = 3.72GTASVVTGVTSTIGLHH214 pKa = 6.9DD215 pKa = 4.0VADD218 pKa = 4.56LDD220 pKa = 4.26VTHH223 pKa = 7.53LDD225 pKa = 3.63PAVSSVVGSVTGSDD239 pKa = 3.34PLGAVSHH246 pKa = 7.3DD247 pKa = 3.51ATQGVLDD254 pKa = 3.79VAGDD258 pKa = 3.56VTSPVTGLVDD268 pKa = 3.72GLGHH272 pKa = 6.01HH273 pKa = 6.73TAPDD277 pKa = 3.97TADD280 pKa = 3.53HH281 pKa = 6.29AHH283 pKa = 6.44GAVTDD288 pKa = 3.99LLFF291 pKa = 4.96
MM1 pKa = 8.03DD2 pKa = 5.63SSPTLQDD9 pKa = 3.27FVLNLIYY16 pKa = 10.8DD17 pKa = 4.28PAARR21 pKa = 11.84SAFEE25 pKa = 4.53LDD27 pKa = 3.62PEE29 pKa = 4.87GVLADD34 pKa = 4.38AGLSDD39 pKa = 3.78VTAVDD44 pKa = 3.73VQEE47 pKa = 4.5VIPLVVDD54 pKa = 3.42YY55 pKa = 11.36APLSDD60 pKa = 3.85VAAVTAPLGVHH71 pKa = 7.1DD72 pKa = 5.36LTTGVADD79 pKa = 4.28ADD81 pKa = 3.7VSAAVAQLQAMTGNLTPGAHH101 pKa = 7.02AAPDD105 pKa = 3.48VAYY108 pKa = 9.95AGAGAFMVSTDD119 pKa = 3.51GLLAGTPAVGVGLEE133 pKa = 4.06HH134 pKa = 7.27AATVDD139 pKa = 3.71APVSTSVTAPVDD151 pKa = 3.64GALSAVHH158 pKa = 7.15DD159 pKa = 4.88PGLGLDD165 pKa = 4.2AGVSTATSVTTTVVGDD181 pKa = 3.86ADD183 pKa = 3.89GLIASTGVDD192 pKa = 3.27AGATLDD198 pKa = 3.72GTASVVTGVTSTIGLHH214 pKa = 6.9DD215 pKa = 4.0VADD218 pKa = 4.56LDD220 pKa = 4.26VTHH223 pKa = 7.53LDD225 pKa = 3.63PAVSSVVGSVTGSDD239 pKa = 3.34PLGAVSHH246 pKa = 7.3DD247 pKa = 3.51ATQGVLDD254 pKa = 3.79VAGDD258 pKa = 3.56VTSPVTGLVDD268 pKa = 3.72GLGHH272 pKa = 6.01HH273 pKa = 6.73TAPDD277 pKa = 3.97TADD280 pKa = 3.53HH281 pKa = 6.29AHH283 pKa = 6.44GAVTDD288 pKa = 3.99LLFF291 pKa = 4.96
Molecular weight: 28.23 kDa
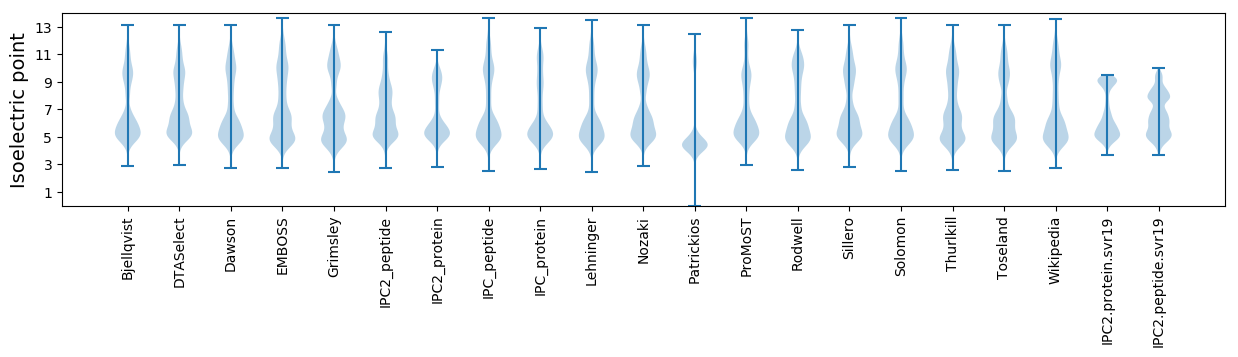
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0SA04|A0A2T0SA04_9ACTN Uncharacterized protein OS=Pseudosporangium ferrugineum OX=439699 GN=CLV70_105381 PE=4 SV=1
MM1 pKa = 7.72PSDD4 pKa = 3.85PTSPSDD10 pKa = 4.23PIRR13 pKa = 11.84STGPRR18 pKa = 11.84RR19 pKa = 11.84CTRR22 pKa = 11.84RR23 pKa = 11.84SGPNRR28 pKa = 11.84RR29 pKa = 11.84ALPRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GPCSGRR44 pKa = 11.84SRR46 pKa = 11.84CTRR49 pKa = 11.84PRR51 pKa = 11.84GPSRR55 pKa = 11.84RR56 pKa = 11.84APRR59 pKa = 11.84SGRR62 pKa = 11.84SPSTRR67 pKa = 11.84HH68 pKa = 6.21RR69 pKa = 11.84GRR71 pKa = 11.84SRR73 pKa = 11.84CSRR76 pKa = 11.84RR77 pKa = 11.84ARR79 pKa = 11.84TRR81 pKa = 11.84PPPRR85 pKa = 11.84SRR87 pKa = 11.84PSRR90 pKa = 11.84NSGTAPRR97 pKa = 11.84HH98 pKa = 4.83PPSRR102 pKa = 11.84RR103 pKa = 11.84PAARR107 pKa = 11.84WTTARR112 pKa = 11.84QRR114 pKa = 11.84SRR116 pKa = 11.84TTARR120 pKa = 11.84HH121 pKa = 5.46AGAPTARR128 pKa = 11.84NPRR131 pKa = 11.84PTPASRR137 pKa = 11.84SHH139 pKa = 5.16SARR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84SRR146 pKa = 11.84SPPARR151 pKa = 11.84SPPSRR156 pKa = 11.84RR157 pKa = 11.84PSLSPAIPSPRR168 pKa = 11.84GLSPPSRR175 pKa = 11.84RR176 pKa = 11.84PSLSPPNPSPPDD188 pKa = 3.35LSPPSRR194 pKa = 11.84RR195 pKa = 11.84VVRR198 pKa = 11.84SRR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 11.84RR203 pKa = 11.84SWSRR207 pKa = 11.84SSRR210 pKa = 11.84RR211 pKa = 11.84LRR213 pKa = 11.84WSARR217 pKa = 11.84PRR219 pKa = 11.84PSPHH223 pKa = 6.64RR224 pKa = 11.84RR225 pKa = 11.84SCAPRR230 pKa = 11.84RR231 pKa = 11.84PSRR234 pKa = 11.84PRR236 pKa = 11.84RR237 pKa = 11.84PPARR241 pKa = 11.84RR242 pKa = 11.84RR243 pKa = 11.84PNRR246 pKa = 11.84PTSARR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84HH255 pKa = 5.27TPTSPSPGTPPPRR268 pKa = 11.84TTAGRR273 pKa = 11.84PLRR276 pKa = 11.84PRR278 pKa = 11.84QRR280 pKa = 11.84WHH282 pKa = 5.81QPLPRR287 pKa = 11.84RR288 pKa = 11.84PHH290 pKa = 6.01PPLWTLRR297 pKa = 11.84PRR299 pKa = 11.84PLRR302 pKa = 11.84RR303 pKa = 11.84HH304 pKa = 5.64RR305 pKa = 11.84RR306 pKa = 11.84PPRR309 pKa = 11.84RR310 pKa = 11.84PRR312 pKa = 11.84SPRR315 pKa = 11.84RR316 pKa = 11.84RR317 pKa = 11.84LHH319 pKa = 6.38RR320 pKa = 11.84RR321 pKa = 11.84RR322 pKa = 11.84LPLRR326 pKa = 11.84HH327 pKa = 5.03QHH329 pKa = 4.5QHH331 pKa = 4.08RR332 pKa = 11.84HH333 pKa = 4.28RR334 pKa = 11.84HH335 pKa = 4.17RR336 pKa = 11.84HH337 pKa = 4.49PLQRR341 pKa = 11.84QRR343 pKa = 11.84PLPRR347 pKa = 11.84RR348 pKa = 11.84CRR350 pKa = 11.84LPRR353 pKa = 11.84RR354 pKa = 11.84RR355 pKa = 11.84PRR357 pKa = 11.84PLPPPRR363 pKa = 11.84LRR365 pKa = 11.84PRR367 pKa = 11.84RR368 pKa = 11.84PPRR371 pKa = 11.84PLLPPRR377 pKa = 11.84LRR379 pKa = 11.84PRR381 pKa = 11.84RR382 pKa = 11.84PQRR385 pKa = 11.84PLPRR389 pKa = 11.84RR390 pKa = 11.84AQRR393 pKa = 11.84PLLPPRR399 pKa = 11.84LRR401 pKa = 11.84PRR403 pKa = 11.84RR404 pKa = 11.84PQRR407 pKa = 11.84PLPRR411 pKa = 11.84RR412 pKa = 11.84RR413 pKa = 11.84PRR415 pKa = 11.84LLPRR419 pKa = 11.84LRR421 pKa = 11.84HH422 pKa = 5.72RR423 pKa = 11.84PLPRR427 pKa = 11.84RR428 pKa = 11.84PPLPHH433 pKa = 6.92RR434 pKa = 11.84RR435 pKa = 11.84PLLRR439 pKa = 11.84RR440 pKa = 11.84RR441 pKa = 11.84CRR443 pKa = 11.84PHH445 pKa = 7.13RR446 pKa = 11.84LLLSHH451 pKa = 7.5RR452 pKa = 11.84SRR454 pKa = 11.84LSRR457 pKa = 11.84RR458 pKa = 11.84RR459 pKa = 11.84RR460 pKa = 11.84LLLSRR465 pKa = 11.84RR466 pKa = 11.84RR467 pKa = 11.84PACPPSRR474 pKa = 11.84RR475 pKa = 11.84KK476 pKa = 10.08RR477 pKa = 11.84PARR480 pKa = 11.84RR481 pKa = 11.84QRR483 pKa = 11.84LRR485 pKa = 11.84LRR487 pKa = 11.84LLRR490 pKa = 11.84FRR492 pKa = 4.94
MM1 pKa = 7.72PSDD4 pKa = 3.85PTSPSDD10 pKa = 4.23PIRR13 pKa = 11.84STGPRR18 pKa = 11.84RR19 pKa = 11.84CTRR22 pKa = 11.84RR23 pKa = 11.84SGPNRR28 pKa = 11.84RR29 pKa = 11.84ALPRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GPCSGRR44 pKa = 11.84SRR46 pKa = 11.84CTRR49 pKa = 11.84PRR51 pKa = 11.84GPSRR55 pKa = 11.84RR56 pKa = 11.84APRR59 pKa = 11.84SGRR62 pKa = 11.84SPSTRR67 pKa = 11.84HH68 pKa = 6.21RR69 pKa = 11.84GRR71 pKa = 11.84SRR73 pKa = 11.84CSRR76 pKa = 11.84RR77 pKa = 11.84ARR79 pKa = 11.84TRR81 pKa = 11.84PPPRR85 pKa = 11.84SRR87 pKa = 11.84PSRR90 pKa = 11.84NSGTAPRR97 pKa = 11.84HH98 pKa = 4.83PPSRR102 pKa = 11.84RR103 pKa = 11.84PAARR107 pKa = 11.84WTTARR112 pKa = 11.84QRR114 pKa = 11.84SRR116 pKa = 11.84TTARR120 pKa = 11.84HH121 pKa = 5.46AGAPTARR128 pKa = 11.84NPRR131 pKa = 11.84PTPASRR137 pKa = 11.84SHH139 pKa = 5.16SARR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84SRR146 pKa = 11.84SPPARR151 pKa = 11.84SPPSRR156 pKa = 11.84RR157 pKa = 11.84PSLSPAIPSPRR168 pKa = 11.84GLSPPSRR175 pKa = 11.84RR176 pKa = 11.84PSLSPPNPSPPDD188 pKa = 3.35LSPPSRR194 pKa = 11.84RR195 pKa = 11.84VVRR198 pKa = 11.84SRR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 11.84RR203 pKa = 11.84SWSRR207 pKa = 11.84SSRR210 pKa = 11.84RR211 pKa = 11.84LRR213 pKa = 11.84WSARR217 pKa = 11.84PRR219 pKa = 11.84PSPHH223 pKa = 6.64RR224 pKa = 11.84RR225 pKa = 11.84SCAPRR230 pKa = 11.84RR231 pKa = 11.84PSRR234 pKa = 11.84PRR236 pKa = 11.84RR237 pKa = 11.84PPARR241 pKa = 11.84RR242 pKa = 11.84RR243 pKa = 11.84PNRR246 pKa = 11.84PTSARR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84HH255 pKa = 5.27TPTSPSPGTPPPRR268 pKa = 11.84TTAGRR273 pKa = 11.84PLRR276 pKa = 11.84PRR278 pKa = 11.84QRR280 pKa = 11.84WHH282 pKa = 5.81QPLPRR287 pKa = 11.84RR288 pKa = 11.84PHH290 pKa = 6.01PPLWTLRR297 pKa = 11.84PRR299 pKa = 11.84PLRR302 pKa = 11.84RR303 pKa = 11.84HH304 pKa = 5.64RR305 pKa = 11.84RR306 pKa = 11.84PPRR309 pKa = 11.84RR310 pKa = 11.84PRR312 pKa = 11.84SPRR315 pKa = 11.84RR316 pKa = 11.84RR317 pKa = 11.84LHH319 pKa = 6.38RR320 pKa = 11.84RR321 pKa = 11.84RR322 pKa = 11.84LPLRR326 pKa = 11.84HH327 pKa = 5.03QHH329 pKa = 4.5QHH331 pKa = 4.08RR332 pKa = 11.84HH333 pKa = 4.28RR334 pKa = 11.84HH335 pKa = 4.17RR336 pKa = 11.84HH337 pKa = 4.49PLQRR341 pKa = 11.84QRR343 pKa = 11.84PLPRR347 pKa = 11.84RR348 pKa = 11.84CRR350 pKa = 11.84LPRR353 pKa = 11.84RR354 pKa = 11.84RR355 pKa = 11.84PRR357 pKa = 11.84PLPPPRR363 pKa = 11.84LRR365 pKa = 11.84PRR367 pKa = 11.84RR368 pKa = 11.84PPRR371 pKa = 11.84PLLPPRR377 pKa = 11.84LRR379 pKa = 11.84PRR381 pKa = 11.84RR382 pKa = 11.84PQRR385 pKa = 11.84PLPRR389 pKa = 11.84RR390 pKa = 11.84AQRR393 pKa = 11.84PLLPPRR399 pKa = 11.84LRR401 pKa = 11.84PRR403 pKa = 11.84RR404 pKa = 11.84PQRR407 pKa = 11.84PLPRR411 pKa = 11.84RR412 pKa = 11.84RR413 pKa = 11.84PRR415 pKa = 11.84LLPRR419 pKa = 11.84LRR421 pKa = 11.84HH422 pKa = 5.72RR423 pKa = 11.84PLPRR427 pKa = 11.84RR428 pKa = 11.84PPLPHH433 pKa = 6.92RR434 pKa = 11.84RR435 pKa = 11.84PLLRR439 pKa = 11.84RR440 pKa = 11.84RR441 pKa = 11.84CRR443 pKa = 11.84PHH445 pKa = 7.13RR446 pKa = 11.84LLLSHH451 pKa = 7.5RR452 pKa = 11.84SRR454 pKa = 11.84LSRR457 pKa = 11.84RR458 pKa = 11.84RR459 pKa = 11.84RR460 pKa = 11.84LLLSRR465 pKa = 11.84RR466 pKa = 11.84RR467 pKa = 11.84PACPPSRR474 pKa = 11.84RR475 pKa = 11.84KK476 pKa = 10.08RR477 pKa = 11.84PARR480 pKa = 11.84RR481 pKa = 11.84QRR483 pKa = 11.84LRR485 pKa = 11.84LRR487 pKa = 11.84LLRR490 pKa = 11.84FRR492 pKa = 4.94
Molecular weight: 58.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2588345 |
28 |
6957 |
334.9 |
35.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.352 ± 0.048 | 0.696 ± 0.008 |
6.0 ± 0.02 | 5.088 ± 0.024 |
2.688 ± 0.017 | 9.544 ± 0.027 |
2.092 ± 0.014 | 3.313 ± 0.019 |
1.772 ± 0.02 | 10.361 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.67 ± 0.01 | 1.784 ± 0.016 |
6.311 ± 0.03 | 2.585 ± 0.016 |
8.388 ± 0.033 | 4.861 ± 0.02 |
6.141 ± 0.024 | 8.77 ± 0.025 |
1.521 ± 0.011 | 2.063 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
