
Formosa sediminum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Formosa
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
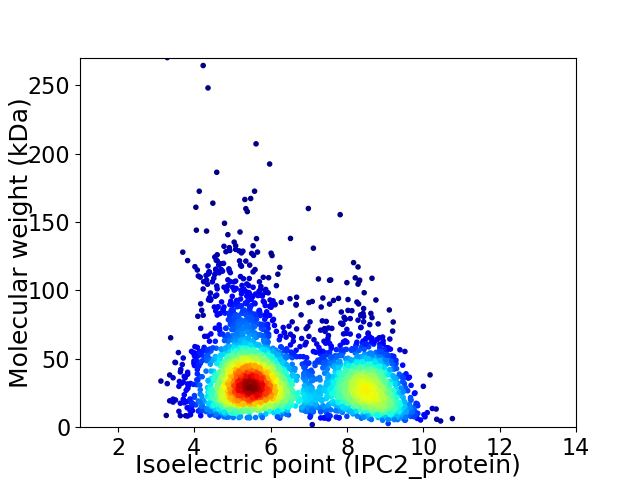
Virtual 2D-PAGE plot for 3315 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
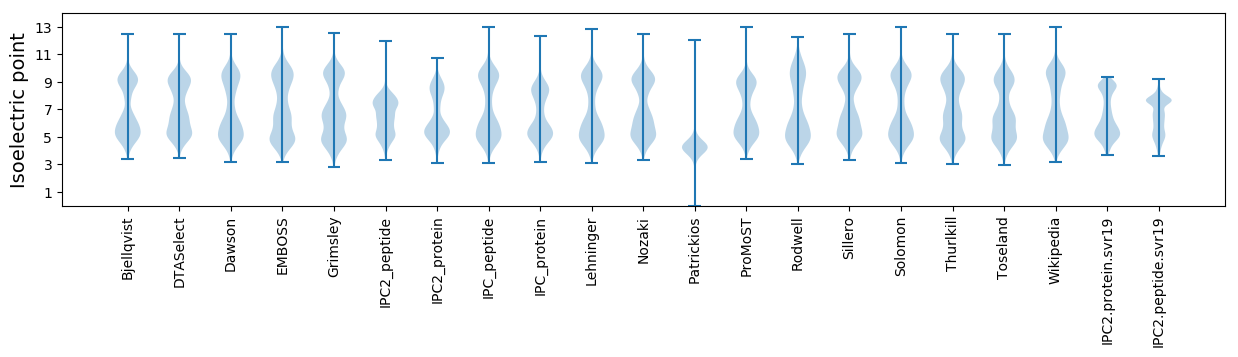
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A516GTL2|A0A516GTL2_9FLAO DUF4907 domain-containing protein OS=Formosa sediminum OX=2594004 GN=FNB79_13015 PE=4 SV=1
MM1 pKa = 7.08ITNSTKK7 pKa = 10.73VLVLCALALSLFNCSSDD24 pKa = 4.88DD25 pKa = 4.75DD26 pKa = 4.65DD27 pKa = 6.43DD28 pKa = 4.71RR29 pKa = 11.84GNWVEE34 pKa = 4.26RR35 pKa = 11.84SVFDD39 pKa = 3.42GVPRR43 pKa = 11.84SNVVGFVIDD52 pKa = 3.47EE53 pKa = 4.31LGYY56 pKa = 10.29MGTGYY61 pKa = 10.86DD62 pKa = 3.69GDD64 pKa = 4.73DD65 pKa = 3.62YY66 pKa = 11.78LVDD69 pKa = 4.15FWQYY73 pKa = 11.52NIAGDD78 pKa = 3.83YY79 pKa = 8.41WVQKK83 pKa = 10.98ADD85 pKa = 3.7FPGTEE90 pKa = 3.83RR91 pKa = 11.84SAATGFALDD100 pKa = 3.45GLGYY104 pKa = 10.69LGTGYY109 pKa = 10.83DD110 pKa = 4.08GVDD113 pKa = 3.42EE114 pKa = 6.33LSDD117 pKa = 3.51FWQYY121 pKa = 11.61NPSTNTWTQKK131 pKa = 10.36ADD133 pKa = 3.32FMGGVRR139 pKa = 11.84QAAIGFGANGTGYY152 pKa = 10.88VGTGYY157 pKa = 10.89DD158 pKa = 3.69GDD160 pKa = 3.92NDD162 pKa = 3.77RR163 pKa = 11.84KK164 pKa = 10.76DD165 pKa = 3.62FYY167 pKa = 11.03KK168 pKa = 11.01YY169 pKa = 11.09NPTTDD174 pKa = 2.84TWSEE178 pKa = 3.74LVGFGGEE185 pKa = 3.76KK186 pKa = 10.02RR187 pKa = 11.84RR188 pKa = 11.84FGTTFAIGDD197 pKa = 3.72KK198 pKa = 10.59VYY200 pKa = 10.69IGTGVSNGLYY210 pKa = 8.68KK211 pKa = 10.35TDD213 pKa = 3.59FWEE216 pKa = 4.35FDD218 pKa = 3.3PATEE222 pKa = 4.25VFTSKK227 pKa = 11.01LDD229 pKa = 3.69LDD231 pKa = 4.33EE232 pKa = 5.29EE233 pKa = 4.89DD234 pKa = 5.06DD235 pKa = 4.18YY236 pKa = 11.99SITRR240 pKa = 11.84SNAVGFSIDD249 pKa = 3.14GLGYY253 pKa = 10.01IVSGYY258 pKa = 10.61NGGVLNSTWEE268 pKa = 3.94YY269 pKa = 11.59DD270 pKa = 3.35PGKK273 pKa = 10.4DD274 pKa = 3.04EE275 pKa = 4.53WEE277 pKa = 5.36DD278 pKa = 3.21ITGLEE283 pKa = 4.74GYY285 pKa = 9.83SRR287 pKa = 11.84QDD289 pKa = 2.94ALAFSTGTRR298 pKa = 11.84AFVLLGRR305 pKa = 11.84SGSLYY310 pKa = 10.73LDD312 pKa = 4.16DD313 pKa = 5.42NYY315 pKa = 11.08EE316 pKa = 4.23LFPQEE321 pKa = 5.74DD322 pKa = 4.3YY323 pKa = 11.62DD324 pKa = 6.31DD325 pKa = 4.29EE326 pKa = 4.69DD327 pKa = 3.54
MM1 pKa = 7.08ITNSTKK7 pKa = 10.73VLVLCALALSLFNCSSDD24 pKa = 4.88DD25 pKa = 4.75DD26 pKa = 4.65DD27 pKa = 6.43DD28 pKa = 4.71RR29 pKa = 11.84GNWVEE34 pKa = 4.26RR35 pKa = 11.84SVFDD39 pKa = 3.42GVPRR43 pKa = 11.84SNVVGFVIDD52 pKa = 3.47EE53 pKa = 4.31LGYY56 pKa = 10.29MGTGYY61 pKa = 10.86DD62 pKa = 3.69GDD64 pKa = 4.73DD65 pKa = 3.62YY66 pKa = 11.78LVDD69 pKa = 4.15FWQYY73 pKa = 11.52NIAGDD78 pKa = 3.83YY79 pKa = 8.41WVQKK83 pKa = 10.98ADD85 pKa = 3.7FPGTEE90 pKa = 3.83RR91 pKa = 11.84SAATGFALDD100 pKa = 3.45GLGYY104 pKa = 10.69LGTGYY109 pKa = 10.83DD110 pKa = 4.08GVDD113 pKa = 3.42EE114 pKa = 6.33LSDD117 pKa = 3.51FWQYY121 pKa = 11.61NPSTNTWTQKK131 pKa = 10.36ADD133 pKa = 3.32FMGGVRR139 pKa = 11.84QAAIGFGANGTGYY152 pKa = 10.88VGTGYY157 pKa = 10.89DD158 pKa = 3.69GDD160 pKa = 3.92NDD162 pKa = 3.77RR163 pKa = 11.84KK164 pKa = 10.76DD165 pKa = 3.62FYY167 pKa = 11.03KK168 pKa = 11.01YY169 pKa = 11.09NPTTDD174 pKa = 2.84TWSEE178 pKa = 3.74LVGFGGEE185 pKa = 3.76KK186 pKa = 10.02RR187 pKa = 11.84RR188 pKa = 11.84FGTTFAIGDD197 pKa = 3.72KK198 pKa = 10.59VYY200 pKa = 10.69IGTGVSNGLYY210 pKa = 8.68KK211 pKa = 10.35TDD213 pKa = 3.59FWEE216 pKa = 4.35FDD218 pKa = 3.3PATEE222 pKa = 4.25VFTSKK227 pKa = 11.01LDD229 pKa = 3.69LDD231 pKa = 4.33EE232 pKa = 5.29EE233 pKa = 4.89DD234 pKa = 5.06DD235 pKa = 4.18YY236 pKa = 11.99SITRR240 pKa = 11.84SNAVGFSIDD249 pKa = 3.14GLGYY253 pKa = 10.01IVSGYY258 pKa = 10.61NGGVLNSTWEE268 pKa = 3.94YY269 pKa = 11.59DD270 pKa = 3.35PGKK273 pKa = 10.4DD274 pKa = 3.04EE275 pKa = 4.53WEE277 pKa = 5.36DD278 pKa = 3.21ITGLEE283 pKa = 4.74GYY285 pKa = 9.83SRR287 pKa = 11.84QDD289 pKa = 2.94ALAFSTGTRR298 pKa = 11.84AFVLLGRR305 pKa = 11.84SGSLYY310 pKa = 10.73LDD312 pKa = 4.16DD313 pKa = 5.42NYY315 pKa = 11.08EE316 pKa = 4.23LFPQEE321 pKa = 5.74DD322 pKa = 4.3YY323 pKa = 11.62DD324 pKa = 6.31DD325 pKa = 4.29EE326 pKa = 4.69DD327 pKa = 3.54
Molecular weight: 36.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A516GRJ6|A0A516GRJ6_9FLAO Inosine-5'-monophosphate dehydrogenase OS=Formosa sediminum OX=2594004 GN=guaB PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1148853 |
14 |
2542 |
346.6 |
39.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.414 ± 0.044 | 0.751 ± 0.013 |
5.673 ± 0.033 | 6.323 ± 0.038 |
5.119 ± 0.034 | 6.148 ± 0.042 |
1.929 ± 0.02 | 8.111 ± 0.04 |
7.599 ± 0.051 | 9.477 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.09 ± 0.019 | 6.244 ± 0.044 |
3.332 ± 0.023 | 3.383 ± 0.023 |
3.164 ± 0.021 | 6.497 ± 0.036 |
6.271 ± 0.041 | 6.197 ± 0.03 |
1.017 ± 0.013 | 4.26 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
