
Giant house spider associated circular virus 3
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.9
Get precalculated fractions of proteins
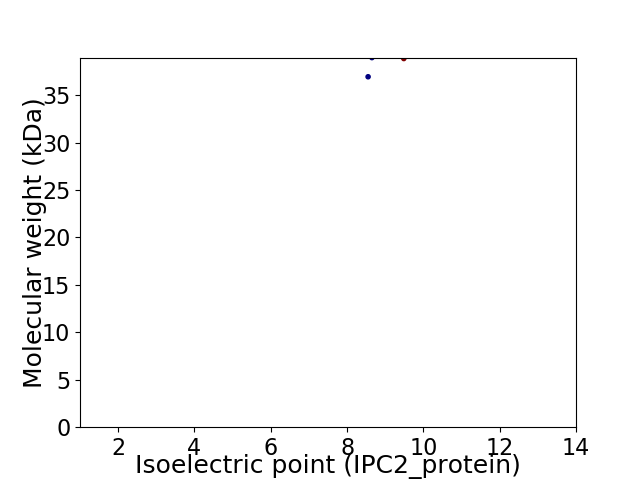
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
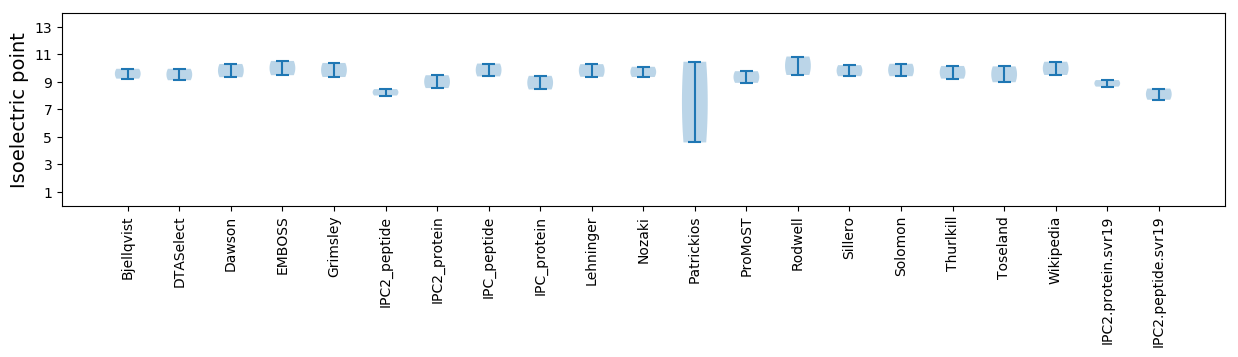
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPD9|A0A346BPD9_9VIRU Putative capsid protein OS=Giant house spider associated circular virus 3 OX=2293290 PE=4 SV=1
MM1 pKa = 7.75ALPDD5 pKa = 3.87KK6 pKa = 10.73RR7 pKa = 11.84NAYY10 pKa = 8.38TPQLLHH16 pKa = 6.7ARR18 pKa = 11.84RR19 pKa = 11.84GSCGEE24 pKa = 3.47QNYY27 pKa = 9.73IKK29 pKa = 10.57RR30 pKa = 11.84PFPRR34 pKa = 11.84FFTMQNEE41 pKa = 4.5LPDD44 pKa = 4.33SKK46 pKa = 10.78PPTVPPSPSSRR57 pKa = 11.84YY58 pKa = 9.66RR59 pKa = 11.84SWCFTKK65 pKa = 10.74NNYY68 pKa = 9.91AISDD72 pKa = 4.11CDD74 pKa = 4.11SILAIPCRR82 pKa = 11.84YY83 pKa = 10.11VIFGKK88 pKa = 10.33EE89 pKa = 3.85VAPTTGTPHH98 pKa = 5.6LQGYY102 pKa = 10.08ISFEE106 pKa = 4.22SAKK109 pKa = 10.51SRR111 pKa = 11.84SAVAKK116 pKa = 9.61LLPGCHH122 pKa = 6.61LLVARR127 pKa = 11.84GTASQNKK134 pKa = 8.28VYY136 pKa = 10.14CSKK139 pKa = 10.95GGQVFEE145 pKa = 4.78NGVLPADD152 pKa = 3.79STVGGRR158 pKa = 11.84MEE160 pKa = 3.85TARR163 pKa = 11.84WDD165 pKa = 4.09NTWSLATSGRR175 pKa = 11.84IEE177 pKa = 5.08EE178 pKa = 4.19IDD180 pKa = 3.06SDD182 pKa = 3.37IRR184 pKa = 11.84LRR186 pKa = 11.84YY187 pKa = 7.41YY188 pKa = 7.98TTIKK192 pKa = 10.3KK193 pKa = 10.14IGVDD197 pKa = 3.19YY198 pKa = 9.63MPPVKK203 pKa = 10.41SLEE206 pKa = 4.43SVCGVWIYY214 pKa = 10.71GASGAGKK221 pKa = 7.84TRR223 pKa = 11.84AVLASFPEE231 pKa = 4.61AYY233 pKa = 9.54IKK235 pKa = 9.97PRR237 pKa = 11.84NVWWDD242 pKa = 3.76GYY244 pKa = 7.13QQEE247 pKa = 4.59EE248 pKa = 5.02VVLLDD253 pKa = 5.04DD254 pKa = 3.91VDD256 pKa = 4.34KK257 pKa = 11.32FDD259 pKa = 3.61VALGGKK265 pKa = 8.96LKK267 pKa = 10.19HH268 pKa = 6.22WADD271 pKa = 4.47FPPFIGEE278 pKa = 3.62IKK280 pKa = 10.42GISRR284 pKa = 11.84RR285 pKa = 11.84IRR287 pKa = 11.84PSKK290 pKa = 10.78LIVTSQYY297 pKa = 11.39KK298 pKa = 8.89MEE300 pKa = 5.34EE301 pKa = 3.83IWNDD305 pKa = 3.29AQTLDD310 pKa = 3.39ALGRR314 pKa = 11.84RR315 pKa = 11.84FKK317 pKa = 11.0VIEE320 pKa = 4.56KK321 pKa = 10.38IKK323 pKa = 10.65GQDD326 pKa = 3.06IILL329 pKa = 4.05
MM1 pKa = 7.75ALPDD5 pKa = 3.87KK6 pKa = 10.73RR7 pKa = 11.84NAYY10 pKa = 8.38TPQLLHH16 pKa = 6.7ARR18 pKa = 11.84RR19 pKa = 11.84GSCGEE24 pKa = 3.47QNYY27 pKa = 9.73IKK29 pKa = 10.57RR30 pKa = 11.84PFPRR34 pKa = 11.84FFTMQNEE41 pKa = 4.5LPDD44 pKa = 4.33SKK46 pKa = 10.78PPTVPPSPSSRR57 pKa = 11.84YY58 pKa = 9.66RR59 pKa = 11.84SWCFTKK65 pKa = 10.74NNYY68 pKa = 9.91AISDD72 pKa = 4.11CDD74 pKa = 4.11SILAIPCRR82 pKa = 11.84YY83 pKa = 10.11VIFGKK88 pKa = 10.33EE89 pKa = 3.85VAPTTGTPHH98 pKa = 5.6LQGYY102 pKa = 10.08ISFEE106 pKa = 4.22SAKK109 pKa = 10.51SRR111 pKa = 11.84SAVAKK116 pKa = 9.61LLPGCHH122 pKa = 6.61LLVARR127 pKa = 11.84GTASQNKK134 pKa = 8.28VYY136 pKa = 10.14CSKK139 pKa = 10.95GGQVFEE145 pKa = 4.78NGVLPADD152 pKa = 3.79STVGGRR158 pKa = 11.84MEE160 pKa = 3.85TARR163 pKa = 11.84WDD165 pKa = 4.09NTWSLATSGRR175 pKa = 11.84IEE177 pKa = 5.08EE178 pKa = 4.19IDD180 pKa = 3.06SDD182 pKa = 3.37IRR184 pKa = 11.84LRR186 pKa = 11.84YY187 pKa = 7.41YY188 pKa = 7.98TTIKK192 pKa = 10.3KK193 pKa = 10.14IGVDD197 pKa = 3.19YY198 pKa = 9.63MPPVKK203 pKa = 10.41SLEE206 pKa = 4.43SVCGVWIYY214 pKa = 10.71GASGAGKK221 pKa = 7.84TRR223 pKa = 11.84AVLASFPEE231 pKa = 4.61AYY233 pKa = 9.54IKK235 pKa = 9.97PRR237 pKa = 11.84NVWWDD242 pKa = 3.76GYY244 pKa = 7.13QQEE247 pKa = 4.59EE248 pKa = 5.02VVLLDD253 pKa = 5.04DD254 pKa = 3.91VDD256 pKa = 4.34KK257 pKa = 11.32FDD259 pKa = 3.61VALGGKK265 pKa = 8.96LKK267 pKa = 10.19HH268 pKa = 6.22WADD271 pKa = 4.47FPPFIGEE278 pKa = 3.62IKK280 pKa = 10.42GISRR284 pKa = 11.84RR285 pKa = 11.84IRR287 pKa = 11.84PSKK290 pKa = 10.78LIVTSQYY297 pKa = 11.39KK298 pKa = 8.89MEE300 pKa = 5.34EE301 pKa = 3.83IWNDD305 pKa = 3.29AQTLDD310 pKa = 3.39ALGRR314 pKa = 11.84RR315 pKa = 11.84FKK317 pKa = 11.0VIEE320 pKa = 4.56KK321 pKa = 10.38IKK323 pKa = 10.65GQDD326 pKa = 3.06IILL329 pKa = 4.05
Molecular weight: 36.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPD9|A0A346BPD9_9VIRU Putative capsid protein OS=Giant house spider associated circular virus 3 OX=2293290 PE=4 SV=1
MM1 pKa = 7.28AAKK4 pKa = 9.83LASAAWRR11 pKa = 11.84NRR13 pKa = 11.84KK14 pKa = 9.43AISAAAAVPAAFFAGRR30 pKa = 11.84KK31 pKa = 6.18NTIKK35 pKa = 10.59RR36 pKa = 11.84KK37 pKa = 8.44ATSQGGRR44 pKa = 11.84PAKK47 pKa = 9.86KK48 pKa = 9.85AKK50 pKa = 7.93QAKK53 pKa = 9.24RR54 pKa = 11.84GTRR57 pKa = 11.84RR58 pKa = 11.84ASSGINVEE66 pKa = 4.2GTINTGKK73 pKa = 10.52KK74 pKa = 8.81IQYY77 pKa = 10.46KK78 pKa = 8.69NVKK81 pKa = 9.2FNKK84 pKa = 9.71SLQLQGGVYY93 pKa = 9.98RR94 pKa = 11.84KK95 pKa = 9.1EE96 pKa = 3.9RR97 pKa = 11.84YY98 pKa = 6.55EE99 pKa = 4.13TGVLYY104 pKa = 10.38HH105 pKa = 7.49DD106 pKa = 5.14PGDD109 pKa = 4.77DD110 pKa = 3.74IFTQKK115 pKa = 10.29VKK117 pKa = 10.15TIDD120 pKa = 3.54FVFDD124 pKa = 3.76GGPPVALGTNITSIFNDD141 pKa = 3.69MYY143 pKa = 11.48ANQATTAPLYY153 pKa = 10.76SSGAVVLNQNEE164 pKa = 3.98ALKK167 pKa = 10.83FFLQSGSIDD176 pKa = 3.75LEE178 pKa = 4.32LTNMSAGSAQLTIYY192 pKa = 11.2VLMAKK197 pKa = 7.78NTKK200 pKa = 8.75STTSGPTADD209 pKa = 3.55WTNGLIAEE217 pKa = 5.1KK218 pKa = 10.25GSQANTPTPFTINNRR233 pKa = 11.84PTSSKK238 pKa = 10.36VFNVNWKK245 pKa = 8.93VCDD248 pKa = 3.6SKK250 pKa = 10.23TYY252 pKa = 10.44NAGPGAKK259 pKa = 9.56VNYY262 pKa = 9.61AFNFSPRR269 pKa = 11.84SVVDD273 pKa = 2.91ATYY276 pKa = 9.81FARR279 pKa = 11.84HH280 pKa = 4.17TQIRR284 pKa = 11.84GLTYY288 pKa = 10.38ALLAVTHH295 pKa = 6.03GQLGLTAAQAVVPKK309 pKa = 9.05PVQWIYY315 pKa = 10.94SIKK318 pKa = 9.21KK319 pKa = 7.68TYY321 pKa = 9.69ILKK324 pKa = 8.82TVNNYY329 pKa = 8.84PAIVSQYY336 pKa = 8.17VQSSATVSASGPVTVRR352 pKa = 11.84EE353 pKa = 4.15DD354 pKa = 3.63DD355 pKa = 4.15GEE357 pKa = 4.57LEE359 pKa = 4.29VV360 pKa = 4.97
MM1 pKa = 7.28AAKK4 pKa = 9.83LASAAWRR11 pKa = 11.84NRR13 pKa = 11.84KK14 pKa = 9.43AISAAAAVPAAFFAGRR30 pKa = 11.84KK31 pKa = 6.18NTIKK35 pKa = 10.59RR36 pKa = 11.84KK37 pKa = 8.44ATSQGGRR44 pKa = 11.84PAKK47 pKa = 9.86KK48 pKa = 9.85AKK50 pKa = 7.93QAKK53 pKa = 9.24RR54 pKa = 11.84GTRR57 pKa = 11.84RR58 pKa = 11.84ASSGINVEE66 pKa = 4.2GTINTGKK73 pKa = 10.52KK74 pKa = 8.81IQYY77 pKa = 10.46KK78 pKa = 8.69NVKK81 pKa = 9.2FNKK84 pKa = 9.71SLQLQGGVYY93 pKa = 9.98RR94 pKa = 11.84KK95 pKa = 9.1EE96 pKa = 3.9RR97 pKa = 11.84YY98 pKa = 6.55EE99 pKa = 4.13TGVLYY104 pKa = 10.38HH105 pKa = 7.49DD106 pKa = 5.14PGDD109 pKa = 4.77DD110 pKa = 3.74IFTQKK115 pKa = 10.29VKK117 pKa = 10.15TIDD120 pKa = 3.54FVFDD124 pKa = 3.76GGPPVALGTNITSIFNDD141 pKa = 3.69MYY143 pKa = 11.48ANQATTAPLYY153 pKa = 10.76SSGAVVLNQNEE164 pKa = 3.98ALKK167 pKa = 10.83FFLQSGSIDD176 pKa = 3.75LEE178 pKa = 4.32LTNMSAGSAQLTIYY192 pKa = 11.2VLMAKK197 pKa = 7.78NTKK200 pKa = 8.75STTSGPTADD209 pKa = 3.55WTNGLIAEE217 pKa = 5.1KK218 pKa = 10.25GSQANTPTPFTINNRR233 pKa = 11.84PTSSKK238 pKa = 10.36VFNVNWKK245 pKa = 8.93VCDD248 pKa = 3.6SKK250 pKa = 10.23TYY252 pKa = 10.44NAGPGAKK259 pKa = 9.56VNYY262 pKa = 9.61AFNFSPRR269 pKa = 11.84SVVDD273 pKa = 2.91ATYY276 pKa = 9.81FARR279 pKa = 11.84HH280 pKa = 4.17TQIRR284 pKa = 11.84GLTYY288 pKa = 10.38ALLAVTHH295 pKa = 6.03GQLGLTAAQAVVPKK309 pKa = 9.05PVQWIYY315 pKa = 10.94SIKK318 pKa = 9.21KK319 pKa = 7.68TYY321 pKa = 9.69ILKK324 pKa = 8.82TVNNYY329 pKa = 8.84PAIVSQYY336 pKa = 8.17VQSSATVSASGPVTVRR352 pKa = 11.84EE353 pKa = 4.15DD354 pKa = 3.63DD355 pKa = 4.15GEE357 pKa = 4.57LEE359 pKa = 4.29VV360 pKa = 4.97
Molecular weight: 38.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
689 |
329 |
360 |
344.5 |
37.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.579 ± 1.803 | 1.161 ± 0.673 |
4.354 ± 0.778 | 3.628 ± 0.86 |
3.774 ± 0.088 | 7.547 ± 0.036 |
1.016 ± 0.139 | 5.951 ± 0.725 |
7.547 ± 0.388 | 6.386 ± 0.421 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.306 ± 0.149 | 4.935 ± 1.32 |
5.515 ± 0.816 | 4.064 ± 0.502 |
5.37 ± 0.917 | 7.547 ± 0.036 |
7.257 ± 1.456 | 7.112 ± 0.508 |
1.742 ± 0.481 | 4.209 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
