
Beihai weivirus-like virus 20
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
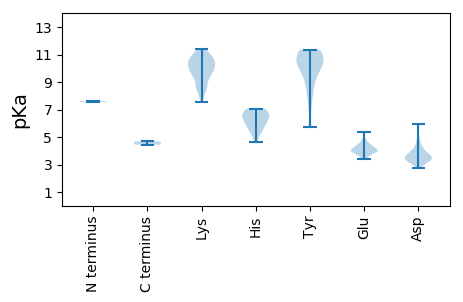
Average proteome isoelectric point is 7.39
Get precalculated fractions of proteins
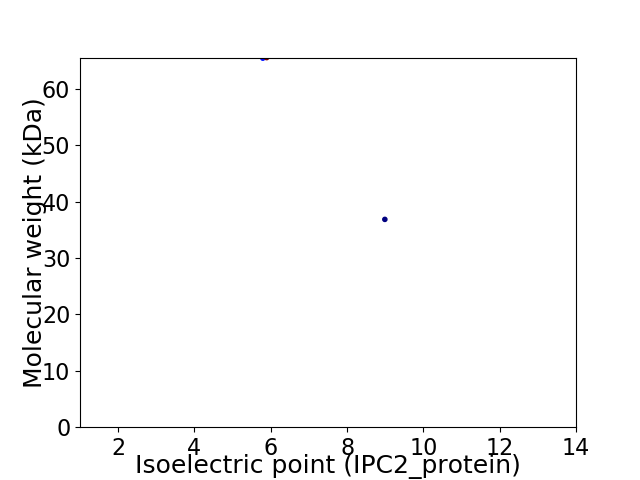
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLD3|A0A1L3KLD3_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 20 OX=1922749 PE=4 SV=1
MM1 pKa = 7.62AEE3 pKa = 3.41AAEE6 pKa = 4.19RR7 pKa = 11.84VNAQQAAFRR16 pKa = 11.84DD17 pKa = 4.23GTTITSTVVEE27 pKa = 4.43APGAASTSGQAEE39 pKa = 4.13GAEE42 pKa = 4.26VGTRR46 pKa = 11.84VARR49 pKa = 11.84PRR51 pKa = 11.84FPSMSEE57 pKa = 3.75EE58 pKa = 4.04EE59 pKa = 4.42VFLFSNDD66 pKa = 3.08PRR68 pKa = 11.84NLEE71 pKa = 3.93AANTLRR77 pKa = 11.84NKK79 pKa = 10.3GVGVHH84 pKa = 5.48QPSEE88 pKa = 4.07KK89 pKa = 9.78EE90 pKa = 3.55AAVRR94 pKa = 11.84DD95 pKa = 3.85EE96 pKa = 5.37LVDD99 pKa = 3.67VLCDD103 pKa = 3.26MVFTKK108 pKa = 10.59RR109 pKa = 11.84EE110 pKa = 4.02SEE112 pKa = 4.16KK113 pKa = 11.39AMMEE117 pKa = 4.17YY118 pKa = 10.75EE119 pKa = 4.5SLTKK123 pKa = 9.8TALPRR128 pKa = 11.84KK129 pKa = 9.72LSEE132 pKa = 4.06EE133 pKa = 4.39QKK135 pKa = 10.93LQWQQDD141 pKa = 3.3ALAAAEE147 pKa = 4.44GDD149 pKa = 3.53GLSYY153 pKa = 11.34SKK155 pKa = 10.91FIDD158 pKa = 3.5AFVKK162 pKa = 10.89AEE164 pKa = 3.97VSAKK168 pKa = 10.0PKK170 pKa = 9.53PRR172 pKa = 11.84PIANHH177 pKa = 5.93KK178 pKa = 9.6EE179 pKa = 3.51IRR181 pKa = 11.84LCALAKK187 pKa = 10.08CAWTYY192 pKa = 8.9EE193 pKa = 4.17HH194 pKa = 7.06VLFRR198 pKa = 11.84RR199 pKa = 11.84MNKK202 pKa = 9.1MSIKK206 pKa = 10.31HH207 pKa = 4.63RR208 pKa = 11.84TKK210 pKa = 10.7SEE212 pKa = 3.91ALSDD216 pKa = 3.71VANALSSMKK225 pKa = 10.39NGRR228 pKa = 11.84WCEE231 pKa = 3.69NDD233 pKa = 2.77LTAFEE238 pKa = 4.84FGISAKK244 pKa = 10.58LKK246 pKa = 9.75ACEE249 pKa = 3.9CAILRR254 pKa = 11.84HH255 pKa = 5.45VASQIGIEE263 pKa = 4.41DD264 pKa = 3.59VGSLLFEE271 pKa = 3.78RR272 pKa = 11.84VVNDD276 pKa = 3.47RR277 pKa = 11.84TKK279 pKa = 8.78TCVWSMRR286 pKa = 11.84YY287 pKa = 7.83TDD289 pKa = 3.38EE290 pKa = 4.31TGEE293 pKa = 3.99KK294 pKa = 9.02RR295 pKa = 11.84TFRR298 pKa = 11.84LILPTVMRR306 pKa = 11.84EE307 pKa = 3.8SGDD310 pKa = 3.56RR311 pKa = 11.84LTSSGNFLQNLIAWCSFLLAPGTVRR336 pKa = 11.84KK337 pKa = 9.9SVEE340 pKa = 3.74SLLRR344 pKa = 11.84TRR346 pKa = 11.84GEE348 pKa = 3.9HH349 pKa = 5.77MFYY352 pKa = 10.88CSARR356 pKa = 11.84DD357 pKa = 3.6GKK359 pKa = 10.95KK360 pKa = 10.31YY361 pKa = 9.81LAKK364 pKa = 10.51LVFEE368 pKa = 5.28GDD370 pKa = 3.58DD371 pKa = 3.31TLGRR375 pKa = 11.84LDD377 pKa = 3.75EE378 pKa = 4.74PVWEE382 pKa = 4.82PKK384 pKa = 10.16RR385 pKa = 11.84EE386 pKa = 3.97GATEE390 pKa = 4.01SLVDD394 pKa = 3.62DD395 pKa = 4.96FFYY398 pKa = 10.79RR399 pKa = 11.84WGWKK403 pKa = 9.42PKK405 pKa = 10.27LSWKK409 pKa = 8.81ATSGYY414 pKa = 10.86DD415 pKa = 3.29YY416 pKa = 11.33ARR418 pKa = 11.84VVGYY422 pKa = 10.35DD423 pKa = 3.2ILIKK427 pKa = 10.89DD428 pKa = 4.11GVAVRR433 pKa = 11.84DD434 pKa = 3.54GDD436 pKa = 4.03KK437 pKa = 10.97LVACPEE443 pKa = 3.76MRR445 pKa = 11.84RR446 pKa = 11.84LLNTKK451 pKa = 8.13QWTTTNVTPEE461 pKa = 3.93EE462 pKa = 4.3LKK464 pKa = 8.58TCNRR468 pKa = 11.84IFAATLAADD477 pKa = 4.67FTRR480 pKa = 11.84VEE482 pKa = 4.01PMYY485 pKa = 11.06SFLRR489 pKa = 11.84SMYY492 pKa = 10.49DD493 pKa = 3.23SNPGGKK499 pKa = 7.54TVTDD503 pKa = 3.56EE504 pKa = 4.02KK505 pKa = 11.0VRR507 pKa = 11.84EE508 pKa = 4.33HH509 pKa = 6.5FLMMTGEE516 pKa = 4.28LPAHH520 pKa = 6.56GSSLLSDD527 pKa = 3.36IAFPEE532 pKa = 3.92FDD534 pKa = 3.25GAGNDD539 pKa = 4.03VEE541 pKa = 4.58WKK543 pKa = 9.67ALARR547 pKa = 11.84VSCGDD552 pKa = 3.49FTDD555 pKa = 5.5LEE557 pKa = 4.46WATACAQPLHH567 pKa = 6.83DD568 pKa = 4.48RR569 pKa = 11.84HH570 pKa = 6.16GADD573 pKa = 4.65LATGMPASWVGTVAA587 pKa = 4.68
MM1 pKa = 7.62AEE3 pKa = 3.41AAEE6 pKa = 4.19RR7 pKa = 11.84VNAQQAAFRR16 pKa = 11.84DD17 pKa = 4.23GTTITSTVVEE27 pKa = 4.43APGAASTSGQAEE39 pKa = 4.13GAEE42 pKa = 4.26VGTRR46 pKa = 11.84VARR49 pKa = 11.84PRR51 pKa = 11.84FPSMSEE57 pKa = 3.75EE58 pKa = 4.04EE59 pKa = 4.42VFLFSNDD66 pKa = 3.08PRR68 pKa = 11.84NLEE71 pKa = 3.93AANTLRR77 pKa = 11.84NKK79 pKa = 10.3GVGVHH84 pKa = 5.48QPSEE88 pKa = 4.07KK89 pKa = 9.78EE90 pKa = 3.55AAVRR94 pKa = 11.84DD95 pKa = 3.85EE96 pKa = 5.37LVDD99 pKa = 3.67VLCDD103 pKa = 3.26MVFTKK108 pKa = 10.59RR109 pKa = 11.84EE110 pKa = 4.02SEE112 pKa = 4.16KK113 pKa = 11.39AMMEE117 pKa = 4.17YY118 pKa = 10.75EE119 pKa = 4.5SLTKK123 pKa = 9.8TALPRR128 pKa = 11.84KK129 pKa = 9.72LSEE132 pKa = 4.06EE133 pKa = 4.39QKK135 pKa = 10.93LQWQQDD141 pKa = 3.3ALAAAEE147 pKa = 4.44GDD149 pKa = 3.53GLSYY153 pKa = 11.34SKK155 pKa = 10.91FIDD158 pKa = 3.5AFVKK162 pKa = 10.89AEE164 pKa = 3.97VSAKK168 pKa = 10.0PKK170 pKa = 9.53PRR172 pKa = 11.84PIANHH177 pKa = 5.93KK178 pKa = 9.6EE179 pKa = 3.51IRR181 pKa = 11.84LCALAKK187 pKa = 10.08CAWTYY192 pKa = 8.9EE193 pKa = 4.17HH194 pKa = 7.06VLFRR198 pKa = 11.84RR199 pKa = 11.84MNKK202 pKa = 9.1MSIKK206 pKa = 10.31HH207 pKa = 4.63RR208 pKa = 11.84TKK210 pKa = 10.7SEE212 pKa = 3.91ALSDD216 pKa = 3.71VANALSSMKK225 pKa = 10.39NGRR228 pKa = 11.84WCEE231 pKa = 3.69NDD233 pKa = 2.77LTAFEE238 pKa = 4.84FGISAKK244 pKa = 10.58LKK246 pKa = 9.75ACEE249 pKa = 3.9CAILRR254 pKa = 11.84HH255 pKa = 5.45VASQIGIEE263 pKa = 4.41DD264 pKa = 3.59VGSLLFEE271 pKa = 3.78RR272 pKa = 11.84VVNDD276 pKa = 3.47RR277 pKa = 11.84TKK279 pKa = 8.78TCVWSMRR286 pKa = 11.84YY287 pKa = 7.83TDD289 pKa = 3.38EE290 pKa = 4.31TGEE293 pKa = 3.99KK294 pKa = 9.02RR295 pKa = 11.84TFRR298 pKa = 11.84LILPTVMRR306 pKa = 11.84EE307 pKa = 3.8SGDD310 pKa = 3.56RR311 pKa = 11.84LTSSGNFLQNLIAWCSFLLAPGTVRR336 pKa = 11.84KK337 pKa = 9.9SVEE340 pKa = 3.74SLLRR344 pKa = 11.84TRR346 pKa = 11.84GEE348 pKa = 3.9HH349 pKa = 5.77MFYY352 pKa = 10.88CSARR356 pKa = 11.84DD357 pKa = 3.6GKK359 pKa = 10.95KK360 pKa = 10.31YY361 pKa = 9.81LAKK364 pKa = 10.51LVFEE368 pKa = 5.28GDD370 pKa = 3.58DD371 pKa = 3.31TLGRR375 pKa = 11.84LDD377 pKa = 3.75EE378 pKa = 4.74PVWEE382 pKa = 4.82PKK384 pKa = 10.16RR385 pKa = 11.84EE386 pKa = 3.97GATEE390 pKa = 4.01SLVDD394 pKa = 3.62DD395 pKa = 4.96FFYY398 pKa = 10.79RR399 pKa = 11.84WGWKK403 pKa = 9.42PKK405 pKa = 10.27LSWKK409 pKa = 8.81ATSGYY414 pKa = 10.86DD415 pKa = 3.29YY416 pKa = 11.33ARR418 pKa = 11.84VVGYY422 pKa = 10.35DD423 pKa = 3.2ILIKK427 pKa = 10.89DD428 pKa = 4.11GVAVRR433 pKa = 11.84DD434 pKa = 3.54GDD436 pKa = 4.03KK437 pKa = 10.97LVACPEE443 pKa = 3.76MRR445 pKa = 11.84RR446 pKa = 11.84LLNTKK451 pKa = 8.13QWTTTNVTPEE461 pKa = 3.93EE462 pKa = 4.3LKK464 pKa = 8.58TCNRR468 pKa = 11.84IFAATLAADD477 pKa = 4.67FTRR480 pKa = 11.84VEE482 pKa = 4.01PMYY485 pKa = 11.06SFLRR489 pKa = 11.84SMYY492 pKa = 10.49DD493 pKa = 3.23SNPGGKK499 pKa = 7.54TVTDD503 pKa = 3.56EE504 pKa = 4.02KK505 pKa = 11.0VRR507 pKa = 11.84EE508 pKa = 4.33HH509 pKa = 6.5FLMMTGEE516 pKa = 4.28LPAHH520 pKa = 6.56GSSLLSDD527 pKa = 3.36IAFPEE532 pKa = 3.92FDD534 pKa = 3.25GAGNDD539 pKa = 4.03VEE541 pKa = 4.58WKK543 pKa = 9.67ALARR547 pKa = 11.84VSCGDD552 pKa = 3.49FTDD555 pKa = 5.5LEE557 pKa = 4.46WATACAQPLHH567 pKa = 6.83DD568 pKa = 4.48RR569 pKa = 11.84HH570 pKa = 6.16GADD573 pKa = 4.65LATGMPASWVGTVAA587 pKa = 4.68
Molecular weight: 65.47 kDa
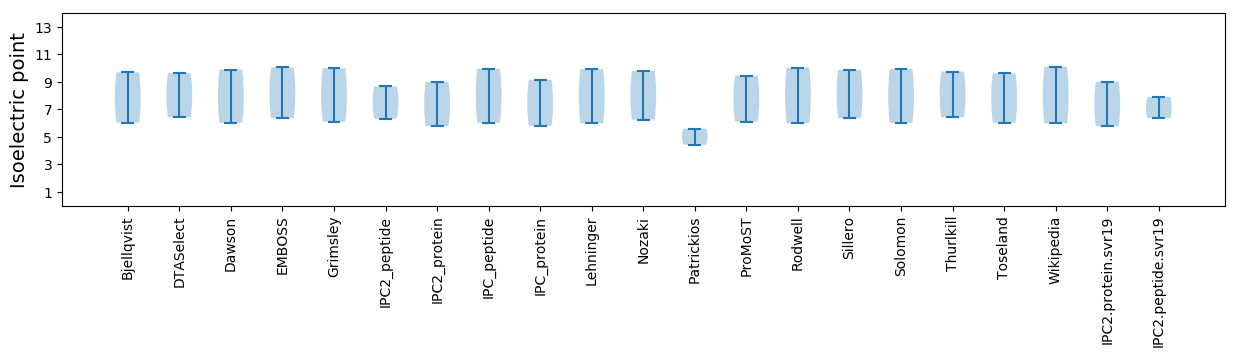
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLD3|A0A1L3KLD3_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 20 OX=1922749 PE=4 SV=1
MM1 pKa = 7.55AGKK4 pKa = 8.56GTRR7 pKa = 11.84KK8 pKa = 9.98NGNNARR14 pKa = 11.84RR15 pKa = 11.84APRR18 pKa = 11.84RR19 pKa = 11.84QGGRR23 pKa = 11.84AANRR27 pKa = 11.84STATRR32 pKa = 11.84VLATGVGASVPKK44 pKa = 10.36PFGSRR49 pKa = 11.84PGQGLACWDD58 pKa = 3.8AKK60 pKa = 10.85HH61 pKa = 6.5PSHH64 pKa = 7.04LALPRR69 pKa = 11.84AVGPYY74 pKa = 5.72TTVRR78 pKa = 11.84ATRR81 pKa = 11.84RR82 pKa = 11.84VQLNTHH88 pKa = 6.44CNVIGTFQEE97 pKa = 4.01GHH99 pKa = 6.54IAPSQAGDD107 pKa = 3.03WSEE110 pKa = 4.14VVLVADD116 pKa = 4.09VTATNPINGHH126 pKa = 6.07LNTRR130 pKa = 11.84SVTVDD135 pKa = 3.22LSGLGDD141 pKa = 3.88AATLVPSALSVQVMCPTALQTASGIIYY168 pKa = 9.82AGVMNTQAALGGRR181 pKa = 11.84VDD183 pKa = 5.98SWDD186 pKa = 3.26DD187 pKa = 3.03WCNKK191 pKa = 8.38FVQFQSPRR199 pKa = 11.84LLAASKK205 pKa = 10.23LALRR209 pKa = 11.84GIQINSYY216 pKa = 8.39PLNMTEE222 pKa = 4.06VSKK225 pKa = 8.46FTPLRR230 pKa = 11.84KK231 pKa = 9.37EE232 pKa = 3.51ADD234 pKa = 3.55LQFEE238 pKa = 4.53YY239 pKa = 10.66TDD241 pKa = 3.36QRR243 pKa = 11.84VEE245 pKa = 3.98PTGWAPIVIYY255 pKa = 10.64NPEE258 pKa = 4.03GTTLEE263 pKa = 4.02LLITVEE269 pKa = 3.69YY270 pKa = 10.11RR271 pKa = 11.84VRR273 pKa = 11.84FDD275 pKa = 4.28LDD277 pKa = 3.4HH278 pKa = 7.02PASASHH284 pKa = 5.36THH286 pKa = 6.25HH287 pKa = 6.96PVASDD292 pKa = 3.69STWDD296 pKa = 3.3RR297 pKa = 11.84MVRR300 pKa = 11.84SATALGNGVMDD311 pKa = 4.02IADD314 pKa = 3.62VVANTGMAVGRR325 pKa = 11.84AAMVGRR331 pKa = 11.84ALTNGAARR339 pKa = 11.84ALPALTMM346 pKa = 4.44
MM1 pKa = 7.55AGKK4 pKa = 8.56GTRR7 pKa = 11.84KK8 pKa = 9.98NGNNARR14 pKa = 11.84RR15 pKa = 11.84APRR18 pKa = 11.84RR19 pKa = 11.84QGGRR23 pKa = 11.84AANRR27 pKa = 11.84STATRR32 pKa = 11.84VLATGVGASVPKK44 pKa = 10.36PFGSRR49 pKa = 11.84PGQGLACWDD58 pKa = 3.8AKK60 pKa = 10.85HH61 pKa = 6.5PSHH64 pKa = 7.04LALPRR69 pKa = 11.84AVGPYY74 pKa = 5.72TTVRR78 pKa = 11.84ATRR81 pKa = 11.84RR82 pKa = 11.84VQLNTHH88 pKa = 6.44CNVIGTFQEE97 pKa = 4.01GHH99 pKa = 6.54IAPSQAGDD107 pKa = 3.03WSEE110 pKa = 4.14VVLVADD116 pKa = 4.09VTATNPINGHH126 pKa = 6.07LNTRR130 pKa = 11.84SVTVDD135 pKa = 3.22LSGLGDD141 pKa = 3.88AATLVPSALSVQVMCPTALQTASGIIYY168 pKa = 9.82AGVMNTQAALGGRR181 pKa = 11.84VDD183 pKa = 5.98SWDD186 pKa = 3.26DD187 pKa = 3.03WCNKK191 pKa = 8.38FVQFQSPRR199 pKa = 11.84LLAASKK205 pKa = 10.23LALRR209 pKa = 11.84GIQINSYY216 pKa = 8.39PLNMTEE222 pKa = 4.06VSKK225 pKa = 8.46FTPLRR230 pKa = 11.84KK231 pKa = 9.37EE232 pKa = 3.51ADD234 pKa = 3.55LQFEE238 pKa = 4.53YY239 pKa = 10.66TDD241 pKa = 3.36QRR243 pKa = 11.84VEE245 pKa = 3.98PTGWAPIVIYY255 pKa = 10.64NPEE258 pKa = 4.03GTTLEE263 pKa = 4.02LLITVEE269 pKa = 3.69YY270 pKa = 10.11RR271 pKa = 11.84VRR273 pKa = 11.84FDD275 pKa = 4.28LDD277 pKa = 3.4HH278 pKa = 7.02PASASHH284 pKa = 5.36THH286 pKa = 6.25HH287 pKa = 6.96PVASDD292 pKa = 3.69STWDD296 pKa = 3.3RR297 pKa = 11.84MVRR300 pKa = 11.84SATALGNGVMDD311 pKa = 4.02IADD314 pKa = 3.62VVANTGMAVGRR325 pKa = 11.84AAMVGRR331 pKa = 11.84ALTNGAARR339 pKa = 11.84ALPALTMM346 pKa = 4.44
Molecular weight: 36.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
933 |
346 |
587 |
466.5 |
51.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.468 ± 0.869 | 1.822 ± 0.376 |
5.466 ± 0.476 | 6.109 ± 1.983 |
3.43 ± 0.795 | 7.181 ± 0.842 |
2.036 ± 0.319 | 2.787 ± 0.222 |
4.716 ± 1.359 | 8.467 ± 0.212 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.787 ± 0.105 | 3.751 ± 0.657 |
4.502 ± 0.723 | 2.68 ± 0.609 |
7.181 ± 0.188 | 6.538 ± 0.265 |
7.503 ± 0.66 | 7.61 ± 0.763 |
2.036 ± 0.171 | 1.929 ± 0.11 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
