
Clostridiales bacterium KLE1615
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; unclassified Eubacteriales
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
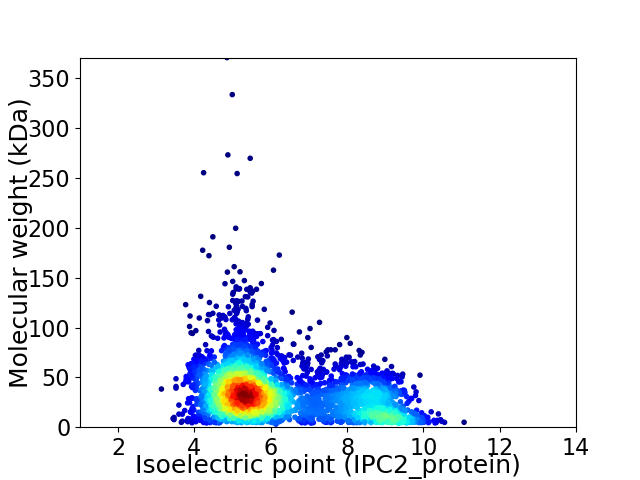
Virtual 2D-PAGE plot for 3669 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A176U395|A0A176U395_9FIRM Histidine kinase OS=Clostridiales bacterium KLE1615 OX=1715004 GN=HMPREF2738_02903 PE=4 SV=1
MM1 pKa = 7.86KK2 pKa = 10.04FRR4 pKa = 11.84KK5 pKa = 9.72ASSLFLAGAMCLSTFGGAMSVFADD29 pKa = 3.78EE30 pKa = 4.58EE31 pKa = 4.64TSSEE35 pKa = 3.98AAEE38 pKa = 4.62AEE40 pKa = 4.41TVDD43 pKa = 4.69YY44 pKa = 11.01SAEE47 pKa = 4.06DD48 pKa = 4.9PITATITVWGPAEE61 pKa = 4.29DD62 pKa = 3.48QAEE65 pKa = 4.49EE66 pKa = 4.07YY67 pKa = 11.25GEE69 pKa = 4.04WLQKK73 pKa = 10.28RR74 pKa = 11.84CEE76 pKa = 4.26AFNEE80 pKa = 4.1LHH82 pKa = 6.64VNWDD86 pKa = 3.33LTFEE90 pKa = 4.33YY91 pKa = 8.26GTCSEE96 pKa = 4.17GDD98 pKa = 3.2AGKK101 pKa = 8.8TISQDD106 pKa = 3.25PSGSADD112 pKa = 2.87VYY114 pKa = 10.87MFANDD119 pKa = 3.6QLQTLIDD126 pKa = 3.7AGAIAEE132 pKa = 4.45LGGKK136 pKa = 6.92TVDD139 pKa = 4.03YY140 pKa = 11.03IKK142 pKa = 10.18DD143 pKa = 3.71TNSEE147 pKa = 4.42AIVDD151 pKa = 4.17SVTVDD156 pKa = 3.22DD157 pKa = 4.82CVYY160 pKa = 10.83GVPFTTNTWYY170 pKa = 9.56MFYY173 pKa = 10.82DD174 pKa = 3.34KK175 pKa = 10.74SVYY178 pKa = 10.47DD179 pKa = 4.24DD180 pKa = 4.57EE181 pKa = 5.44DD182 pKa = 3.96VKK184 pKa = 11.61SLDD187 pKa = 3.34TMLEE191 pKa = 3.87KK192 pKa = 10.98GKK194 pKa = 10.98VSFPLTNSWYY204 pKa = 9.39IASFYY209 pKa = 10.59VGNGCTLFGEE219 pKa = 4.79DD220 pKa = 4.21GTDD223 pKa = 3.24EE224 pKa = 3.91EE225 pKa = 5.41AGIDD229 pKa = 3.69FAGEE233 pKa = 3.39KK234 pKa = 9.87GAAVTKK240 pKa = 10.49YY241 pKa = 10.42LVNLVGNKK249 pKa = 9.84NFVNDD254 pKa = 3.54EE255 pKa = 4.38SGVGVSGMCDD265 pKa = 3.09GSINAMFSGSWDD277 pKa = 3.49YY278 pKa = 11.55TKK280 pKa = 10.79LSEE283 pKa = 5.42AMGDD287 pKa = 3.25NLGIVKK293 pKa = 10.51LPTYY297 pKa = 10.42NLDD300 pKa = 3.45GEE302 pKa = 4.7EE303 pKa = 3.91IQMEE307 pKa = 4.4SFAGSKK313 pKa = 10.57AIGVNPNCEE322 pKa = 4.07YY323 pKa = 10.39PQVAVALALFLGNEE337 pKa = 4.62EE338 pKa = 4.36SQQMHH343 pKa = 6.47YY344 pKa = 10.18DD345 pKa = 3.02ARR347 pKa = 11.84SIVPCNTEE355 pKa = 3.79LLEE358 pKa = 4.35EE359 pKa = 4.2EE360 pKa = 5.21NIQKK364 pKa = 10.68DD365 pKa = 4.08EE366 pKa = 4.13LVIAQNEE373 pKa = 4.62TFDD376 pKa = 4.01EE377 pKa = 4.49TSILQPFVSAMNNYY391 pKa = 6.51WTPAEE396 pKa = 4.19NFGKK400 pKa = 10.48SIVNGEE406 pKa = 4.11VTLDD410 pKa = 3.42NAEE413 pKa = 3.98EE414 pKa = 4.15MTDD417 pKa = 3.5KK418 pKa = 11.22LSDD421 pKa = 3.53SMNQSIVDD429 pKa = 3.48
MM1 pKa = 7.86KK2 pKa = 10.04FRR4 pKa = 11.84KK5 pKa = 9.72ASSLFLAGAMCLSTFGGAMSVFADD29 pKa = 3.78EE30 pKa = 4.58EE31 pKa = 4.64TSSEE35 pKa = 3.98AAEE38 pKa = 4.62AEE40 pKa = 4.41TVDD43 pKa = 4.69YY44 pKa = 11.01SAEE47 pKa = 4.06DD48 pKa = 4.9PITATITVWGPAEE61 pKa = 4.29DD62 pKa = 3.48QAEE65 pKa = 4.49EE66 pKa = 4.07YY67 pKa = 11.25GEE69 pKa = 4.04WLQKK73 pKa = 10.28RR74 pKa = 11.84CEE76 pKa = 4.26AFNEE80 pKa = 4.1LHH82 pKa = 6.64VNWDD86 pKa = 3.33LTFEE90 pKa = 4.33YY91 pKa = 8.26GTCSEE96 pKa = 4.17GDD98 pKa = 3.2AGKK101 pKa = 8.8TISQDD106 pKa = 3.25PSGSADD112 pKa = 2.87VYY114 pKa = 10.87MFANDD119 pKa = 3.6QLQTLIDD126 pKa = 3.7AGAIAEE132 pKa = 4.45LGGKK136 pKa = 6.92TVDD139 pKa = 4.03YY140 pKa = 11.03IKK142 pKa = 10.18DD143 pKa = 3.71TNSEE147 pKa = 4.42AIVDD151 pKa = 4.17SVTVDD156 pKa = 3.22DD157 pKa = 4.82CVYY160 pKa = 10.83GVPFTTNTWYY170 pKa = 9.56MFYY173 pKa = 10.82DD174 pKa = 3.34KK175 pKa = 10.74SVYY178 pKa = 10.47DD179 pKa = 4.24DD180 pKa = 4.57EE181 pKa = 5.44DD182 pKa = 3.96VKK184 pKa = 11.61SLDD187 pKa = 3.34TMLEE191 pKa = 3.87KK192 pKa = 10.98GKK194 pKa = 10.98VSFPLTNSWYY204 pKa = 9.39IASFYY209 pKa = 10.59VGNGCTLFGEE219 pKa = 4.79DD220 pKa = 4.21GTDD223 pKa = 3.24EE224 pKa = 3.91EE225 pKa = 5.41AGIDD229 pKa = 3.69FAGEE233 pKa = 3.39KK234 pKa = 9.87GAAVTKK240 pKa = 10.49YY241 pKa = 10.42LVNLVGNKK249 pKa = 9.84NFVNDD254 pKa = 3.54EE255 pKa = 4.38SGVGVSGMCDD265 pKa = 3.09GSINAMFSGSWDD277 pKa = 3.49YY278 pKa = 11.55TKK280 pKa = 10.79LSEE283 pKa = 5.42AMGDD287 pKa = 3.25NLGIVKK293 pKa = 10.51LPTYY297 pKa = 10.42NLDD300 pKa = 3.45GEE302 pKa = 4.7EE303 pKa = 3.91IQMEE307 pKa = 4.4SFAGSKK313 pKa = 10.57AIGVNPNCEE322 pKa = 4.07YY323 pKa = 10.39PQVAVALALFLGNEE337 pKa = 4.62EE338 pKa = 4.36SQQMHH343 pKa = 6.47YY344 pKa = 10.18DD345 pKa = 3.02ARR347 pKa = 11.84SIVPCNTEE355 pKa = 3.79LLEE358 pKa = 4.35EE359 pKa = 4.2EE360 pKa = 5.21NIQKK364 pKa = 10.68DD365 pKa = 4.08EE366 pKa = 4.13LVIAQNEE373 pKa = 4.62TFDD376 pKa = 4.01EE377 pKa = 4.49TSILQPFVSAMNNYY391 pKa = 6.51WTPAEE396 pKa = 4.19NFGKK400 pKa = 10.48SIVNGEE406 pKa = 4.11VTLDD410 pKa = 3.42NAEE413 pKa = 3.98EE414 pKa = 4.15MTDD417 pKa = 3.5KK418 pKa = 11.22LSDD421 pKa = 3.53SMNQSIVDD429 pKa = 3.48
Molecular weight: 46.81 kDa
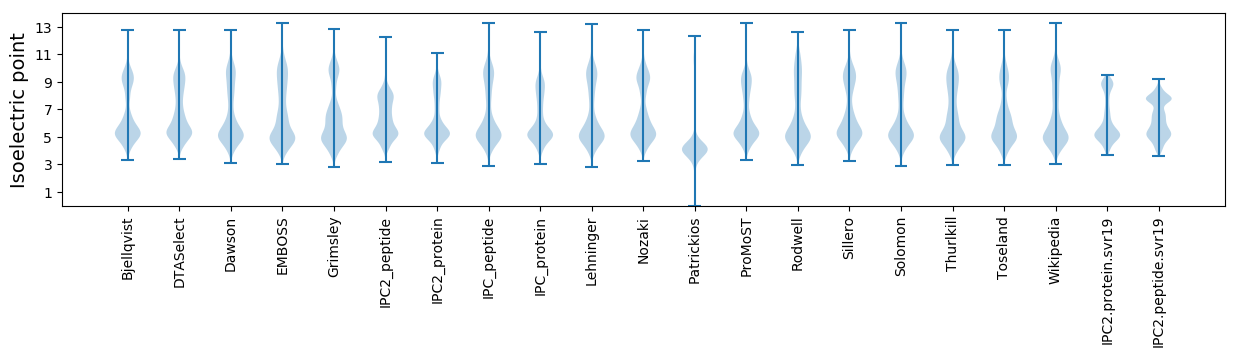
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A176U712|A0A176U712_9FIRM Beta-glucosidase OS=Clostridiales bacterium KLE1615 OX=1715004 GN=HMPREF2738_01404 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.3KK9 pKa = 8.13RR10 pKa = 11.84SHH12 pKa = 6.3AKK14 pKa = 8.8VHH16 pKa = 5.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.78VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.3KK9 pKa = 8.13RR10 pKa = 11.84SHH12 pKa = 6.3AKK14 pKa = 8.8VHH16 pKa = 5.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.78VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 4.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1184311 |
27 |
3472 |
322.8 |
36.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.565 ± 0.042 | 1.612 ± 0.017 |
5.674 ± 0.032 | 7.438 ± 0.049 |
4.183 ± 0.026 | 6.637 ± 0.037 |
1.821 ± 0.017 | 7.033 ± 0.04 |
6.735 ± 0.035 | 8.883 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.135 ± 0.023 | 4.38 ± 0.027 |
3.288 ± 0.025 | 3.615 ± 0.027 |
4.292 ± 0.033 | 6.062 ± 0.037 |
5.575 ± 0.04 | 6.624 ± 0.033 |
1.146 ± 0.017 | 4.304 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
