
Mycobacterium sp. 1274756.6
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium; unclassified Mycobacterium
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
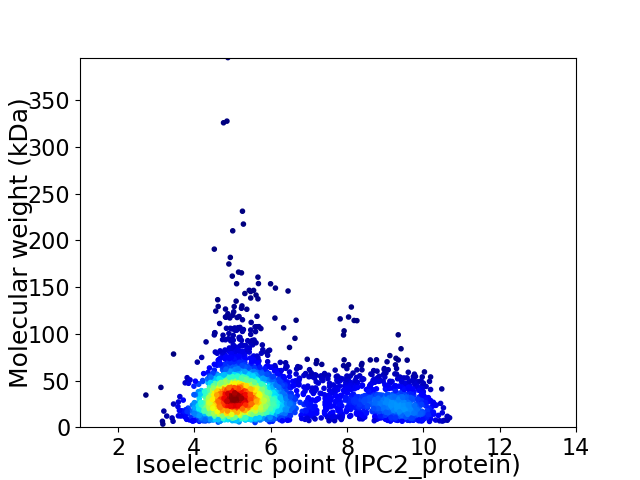
Virtual 2D-PAGE plot for 3346 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A3JNC1|A0A1A3JNC1_9MYCO Rieske domain-containing protein OS=Mycobacterium sp. 1274756.6 OX=1834076 GN=A5643_02830 PE=4 SV=1
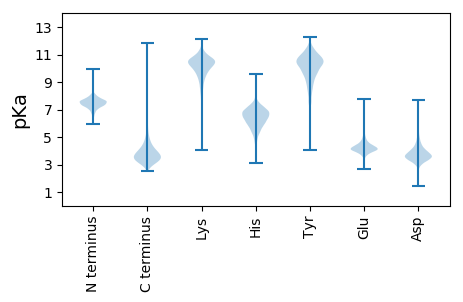
MM1 pKa = 7.17NPVHH5 pKa = 6.67RR6 pKa = 11.84RR7 pKa = 11.84AGAALTALTITAAGLLGLPPVSPLLDD33 pKa = 3.3LTTFAVHH40 pKa = 5.89LTADD44 pKa = 3.37ADD46 pKa = 4.06ADD48 pKa = 4.21VVAVPPALVMGGSGVPNPSAGWVDD72 pKa = 3.67SASRR76 pKa = 11.84LYY78 pKa = 10.33LQPLGFGGEE87 pKa = 3.99AQSFYY92 pKa = 10.76TPEE95 pKa = 4.4LMGQHH100 pKa = 7.19PDD102 pKa = 2.31WSMAEE107 pKa = 4.25GARR110 pKa = 11.84LLTAGIHH117 pKa = 6.41ANLATASAEE126 pKa = 3.85NPVYY130 pKa = 10.95VFGYY134 pKa = 8.55SQSSALSTLTMNQLAADD151 pKa = 4.41GVDD154 pKa = 2.93QDD156 pKa = 4.57LVHH159 pKa = 6.72FVLVGNSANPNGGMLTSFDD178 pKa = 4.26LPLIRR183 pKa = 11.84DD184 pKa = 3.9ISRR187 pKa = 11.84MFEE190 pKa = 3.86MTLGNPTPNLFPTDD204 pKa = 3.91VFTLEE209 pKa = 3.76YY210 pKa = 10.46DD211 pKa = 4.18GYY213 pKa = 11.47ADD215 pKa = 4.69FPRR218 pKa = 11.84YY219 pKa = 8.78PLNPLSTLNALVGLFTQHH237 pKa = 6.41LKK239 pKa = 10.97YY240 pKa = 10.72LGFGYY245 pKa = 10.58DD246 pKa = 4.83DD247 pKa = 4.21LWSSPGVVNPANVVQLDD264 pKa = 4.07TVGDD268 pKa = 3.68LTDD271 pKa = 3.64YY272 pKa = 11.14YY273 pKa = 10.24IIRR276 pKa = 11.84ADD278 pKa = 4.05TLPLLDD284 pKa = 4.63PLRR287 pKa = 11.84LVPVAGRR294 pKa = 11.84PLADD298 pKa = 4.2LLEE301 pKa = 4.91PDD303 pKa = 4.94LRR305 pKa = 11.84ILVNLGYY312 pKa = 11.05GSITDD317 pKa = 3.83GWNSGPADD325 pKa = 3.56VASTIGLFPTEE336 pKa = 3.98LHH338 pKa = 6.5CVAGCSWLDD347 pKa = 3.5LPEE350 pKa = 5.32ALLRR354 pKa = 11.84GAWQGVSDD362 pKa = 5.11FVDD365 pKa = 4.2DD366 pKa = 5.2LLDD369 pKa = 3.74PTTYY373 pKa = 10.96QLADD377 pKa = 5.11PITSPALTTLVNAAINTGVVDD398 pKa = 4.24SAPSSLWEE406 pKa = 4.76LLSAWLDD413 pKa = 3.76TSATPLDD420 pKa = 4.26FGALAGQLGLGGLLDD435 pKa = 3.71WVTTLLASLTGG446 pKa = 3.58
MM1 pKa = 7.17NPVHH5 pKa = 6.67RR6 pKa = 11.84RR7 pKa = 11.84AGAALTALTITAAGLLGLPPVSPLLDD33 pKa = 3.3LTTFAVHH40 pKa = 5.89LTADD44 pKa = 3.37ADD46 pKa = 4.06ADD48 pKa = 4.21VVAVPPALVMGGSGVPNPSAGWVDD72 pKa = 3.67SASRR76 pKa = 11.84LYY78 pKa = 10.33LQPLGFGGEE87 pKa = 3.99AQSFYY92 pKa = 10.76TPEE95 pKa = 4.4LMGQHH100 pKa = 7.19PDD102 pKa = 2.31WSMAEE107 pKa = 4.25GARR110 pKa = 11.84LLTAGIHH117 pKa = 6.41ANLATASAEE126 pKa = 3.85NPVYY130 pKa = 10.95VFGYY134 pKa = 8.55SQSSALSTLTMNQLAADD151 pKa = 4.41GVDD154 pKa = 2.93QDD156 pKa = 4.57LVHH159 pKa = 6.72FVLVGNSANPNGGMLTSFDD178 pKa = 4.26LPLIRR183 pKa = 11.84DD184 pKa = 3.9ISRR187 pKa = 11.84MFEE190 pKa = 3.86MTLGNPTPNLFPTDD204 pKa = 3.91VFTLEE209 pKa = 3.76YY210 pKa = 10.46DD211 pKa = 4.18GYY213 pKa = 11.47ADD215 pKa = 4.69FPRR218 pKa = 11.84YY219 pKa = 8.78PLNPLSTLNALVGLFTQHH237 pKa = 6.41LKK239 pKa = 10.97YY240 pKa = 10.72LGFGYY245 pKa = 10.58DD246 pKa = 4.83DD247 pKa = 4.21LWSSPGVVNPANVVQLDD264 pKa = 4.07TVGDD268 pKa = 3.68LTDD271 pKa = 3.64YY272 pKa = 11.14YY273 pKa = 10.24IIRR276 pKa = 11.84ADD278 pKa = 4.05TLPLLDD284 pKa = 4.63PLRR287 pKa = 11.84LVPVAGRR294 pKa = 11.84PLADD298 pKa = 4.2LLEE301 pKa = 4.91PDD303 pKa = 4.94LRR305 pKa = 11.84ILVNLGYY312 pKa = 11.05GSITDD317 pKa = 3.83GWNSGPADD325 pKa = 3.56VASTIGLFPTEE336 pKa = 3.98LHH338 pKa = 6.5CVAGCSWLDD347 pKa = 3.5LPEE350 pKa = 5.32ALLRR354 pKa = 11.84GAWQGVSDD362 pKa = 5.11FVDD365 pKa = 4.2DD366 pKa = 5.2LLDD369 pKa = 3.74PTTYY373 pKa = 10.96QLADD377 pKa = 5.11PITSPALTTLVNAAINTGVVDD398 pKa = 4.24SAPSSLWEE406 pKa = 4.76LLSAWLDD413 pKa = 3.76TSATPLDD420 pKa = 4.26FGALAGQLGLGGLLDD435 pKa = 3.71WVTTLLASLTGG446 pKa = 3.58
Molecular weight: 47.03 kDa
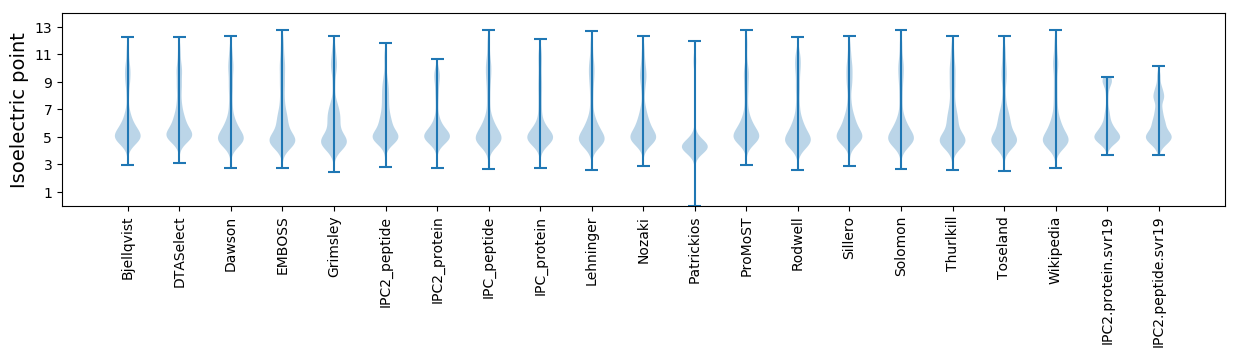
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A3JM91|A0A1A3JM91_9MYCO Mycobactin synthetase protein B OS=Mycobacterium sp. 1274756.6 OX=1834076 GN=A5643_04955 PE=3 SV=1
MM1 pKa = 7.56LRR3 pKa = 11.84RR4 pKa = 11.84LSALPIDD11 pKa = 4.45KK12 pKa = 10.52FLLALLATVAVATVLPASGAAADD35 pKa = 3.93VLEE38 pKa = 4.62VATKK42 pKa = 10.51AAIALLFWLYY52 pKa = 8.77GTRR55 pKa = 11.84LSPRR59 pKa = 11.84QAWQGLRR66 pKa = 11.84HH67 pKa = 5.48WQLHH71 pKa = 5.52LLVLAITFVVFPVLGLIARR90 pKa = 11.84SLVPTVLTIDD100 pKa = 4.87LYY102 pKa = 10.83TGLLFLCLVPSTVQSSIAFTSIARR126 pKa = 11.84GNVSAAIVSASVSNIVGVVVTPLLVMLLMHH156 pKa = 6.79TGGVPGMDD164 pKa = 3.23PTAIRR169 pKa = 11.84DD170 pKa = 3.6IVVQLLLPFLAGQLMRR186 pKa = 11.84PWVADD191 pKa = 4.48WIARR195 pKa = 11.84HH196 pKa = 5.58AALTRR201 pKa = 11.84VVDD204 pKa = 3.95RR205 pKa = 11.84GSILLVVYY213 pKa = 7.89TAFSIGMTRR222 pKa = 11.84HH223 pKa = 5.07IWSTVAPWQLVAVTVVSALLLALVMGITVVAGQLADD259 pKa = 4.25FSRR262 pKa = 11.84ADD264 pKa = 4.2RR265 pKa = 11.84IVLLFCGSKK274 pKa = 10.38KK275 pKa = 10.6SLASGLPMALVLFPAATVGITMLPLMLFHH304 pKa = 6.9QIQLIVCAVIAARR317 pKa = 11.84LARR320 pKa = 11.84TADD323 pKa = 3.5AAADD327 pKa = 3.73PAA329 pKa = 4.69
MM1 pKa = 7.56LRR3 pKa = 11.84RR4 pKa = 11.84LSALPIDD11 pKa = 4.45KK12 pKa = 10.52FLLALLATVAVATVLPASGAAADD35 pKa = 3.93VLEE38 pKa = 4.62VATKK42 pKa = 10.51AAIALLFWLYY52 pKa = 8.77GTRR55 pKa = 11.84LSPRR59 pKa = 11.84QAWQGLRR66 pKa = 11.84HH67 pKa = 5.48WQLHH71 pKa = 5.52LLVLAITFVVFPVLGLIARR90 pKa = 11.84SLVPTVLTIDD100 pKa = 4.87LYY102 pKa = 10.83TGLLFLCLVPSTVQSSIAFTSIARR126 pKa = 11.84GNVSAAIVSASVSNIVGVVVTPLLVMLLMHH156 pKa = 6.79TGGVPGMDD164 pKa = 3.23PTAIRR169 pKa = 11.84DD170 pKa = 3.6IVVQLLLPFLAGQLMRR186 pKa = 11.84PWVADD191 pKa = 4.48WIARR195 pKa = 11.84HH196 pKa = 5.58AALTRR201 pKa = 11.84VVDD204 pKa = 3.95RR205 pKa = 11.84GSILLVVYY213 pKa = 7.89TAFSIGMTRR222 pKa = 11.84HH223 pKa = 5.07IWSTVAPWQLVAVTVVSALLLALVMGITVVAGQLADD259 pKa = 4.25FSRR262 pKa = 11.84ADD264 pKa = 4.2RR265 pKa = 11.84IVLLFCGSKK274 pKa = 10.38KK275 pKa = 10.6SLASGLPMALVLFPAATVGITMLPLMLFHH304 pKa = 6.9QIQLIVCAVIAARR317 pKa = 11.84LARR320 pKa = 11.84TADD323 pKa = 3.5AAADD327 pKa = 3.73PAA329 pKa = 4.69
Molecular weight: 34.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1103034 |
30 |
3722 |
329.7 |
35.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.07 ± 0.073 | 0.762 ± 0.012 |
6.399 ± 0.031 | 5.24 ± 0.041 |
2.92 ± 0.025 | 8.921 ± 0.034 |
2.163 ± 0.018 | 3.937 ± 0.03 |
1.926 ± 0.028 | 10.126 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.889 ± 0.018 | 2.0 ± 0.021 |
6.042 ± 0.039 | 2.945 ± 0.021 |
7.601 ± 0.042 | 4.748 ± 0.027 |
6.092 ± 0.024 | 8.648 ± 0.039 |
1.468 ± 0.017 | 2.103 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
