
Raspberry leaf blotch emaravirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Fimoviridae; Emaravirus
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
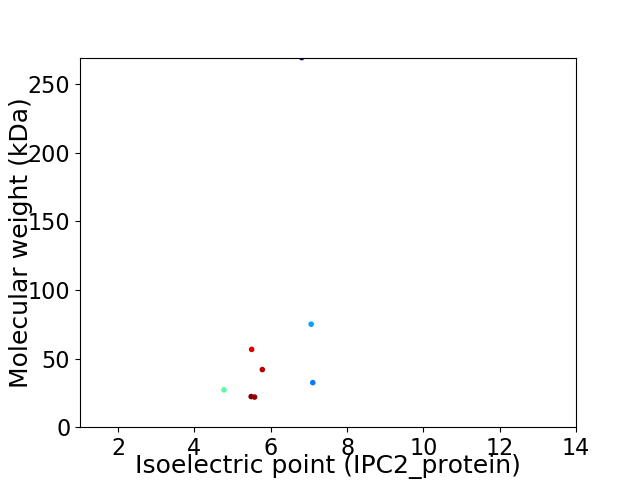
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8K9Y7|F8K9Y7_9VIRU Replicase OS=Raspberry leaf blotch emaravirus OX=1980431 GN=RDRP PE=4 SV=1
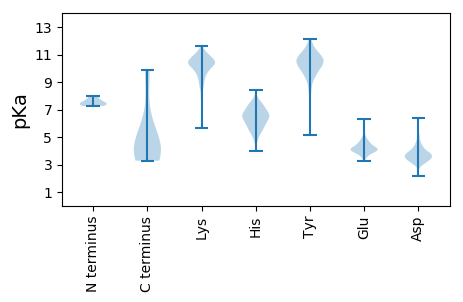
MM1 pKa = 7.99SEE3 pKa = 3.48IGITIEE9 pKa = 6.14DD10 pKa = 3.52ILEE13 pKa = 4.18NLWIEE18 pKa = 4.16LFNAISEE25 pKa = 4.36YY26 pKa = 11.27AKK28 pKa = 10.72VGEE31 pKa = 4.14YY32 pKa = 10.81DD33 pKa = 4.37DD34 pKa = 6.23LINLKK39 pKa = 10.73DD40 pKa = 3.66EE41 pKa = 4.67LLDD44 pKa = 4.01GGEE47 pKa = 4.53AEE49 pKa = 4.78SFSKK53 pKa = 10.73PGLKK57 pKa = 10.49VIFKK61 pKa = 9.88FNKK64 pKa = 9.49LAFICTQIVDD74 pKa = 4.06YY75 pKa = 11.57DD76 pKa = 3.88NILYY80 pKa = 10.42RR81 pKa = 11.84KK82 pKa = 9.25FKK84 pKa = 10.75NHH86 pKa = 6.3FDD88 pKa = 3.27KK89 pKa = 11.36FGIKK93 pKa = 8.88MLNKK97 pKa = 10.25DD98 pKa = 3.06YY99 pKa = 10.15TLRR102 pKa = 11.84YY103 pKa = 9.29RR104 pKa = 11.84NPIGLARR111 pKa = 11.84EE112 pKa = 4.16IFNHH116 pKa = 5.56VNFKK120 pKa = 10.05MDD122 pKa = 3.93LSDD125 pKa = 3.45VEE127 pKa = 4.3YY128 pKa = 10.4WNGFFITITGEE139 pKa = 3.95LNKK142 pKa = 9.69TKK144 pKa = 10.57SIMEE148 pKa = 3.76GDD150 pKa = 3.71YY151 pKa = 10.59IVRR154 pKa = 11.84CEE156 pKa = 3.89NQLKK160 pKa = 10.17IIHH163 pKa = 6.64KK164 pKa = 9.71LDD166 pKa = 4.42GIIRR170 pKa = 11.84TIQKK174 pKa = 9.94KK175 pKa = 9.73SKK177 pKa = 9.55EE178 pKa = 4.15YY179 pKa = 10.67DD180 pKa = 2.94SSTIKK185 pKa = 10.85NYY187 pKa = 9.84IDD189 pKa = 3.06FTMYY193 pKa = 11.0LDD195 pKa = 3.97DD196 pKa = 4.28CSYY199 pKa = 11.28VVSSSQVRR207 pKa = 11.84RR208 pKa = 11.84NDD210 pKa = 3.09EE211 pKa = 4.16DD212 pKa = 3.76CAEE215 pKa = 4.16TVIDD219 pKa = 4.3EE220 pKa = 4.62NQDD223 pKa = 3.31KK224 pKa = 11.33LNIAEE229 pKa = 4.3SDD231 pKa = 3.48ATT233 pKa = 3.76
MM1 pKa = 7.99SEE3 pKa = 3.48IGITIEE9 pKa = 6.14DD10 pKa = 3.52ILEE13 pKa = 4.18NLWIEE18 pKa = 4.16LFNAISEE25 pKa = 4.36YY26 pKa = 11.27AKK28 pKa = 10.72VGEE31 pKa = 4.14YY32 pKa = 10.81DD33 pKa = 4.37DD34 pKa = 6.23LINLKK39 pKa = 10.73DD40 pKa = 3.66EE41 pKa = 4.67LLDD44 pKa = 4.01GGEE47 pKa = 4.53AEE49 pKa = 4.78SFSKK53 pKa = 10.73PGLKK57 pKa = 10.49VIFKK61 pKa = 9.88FNKK64 pKa = 9.49LAFICTQIVDD74 pKa = 4.06YY75 pKa = 11.57DD76 pKa = 3.88NILYY80 pKa = 10.42RR81 pKa = 11.84KK82 pKa = 9.25FKK84 pKa = 10.75NHH86 pKa = 6.3FDD88 pKa = 3.27KK89 pKa = 11.36FGIKK93 pKa = 8.88MLNKK97 pKa = 10.25DD98 pKa = 3.06YY99 pKa = 10.15TLRR102 pKa = 11.84YY103 pKa = 9.29RR104 pKa = 11.84NPIGLARR111 pKa = 11.84EE112 pKa = 4.16IFNHH116 pKa = 5.56VNFKK120 pKa = 10.05MDD122 pKa = 3.93LSDD125 pKa = 3.45VEE127 pKa = 4.3YY128 pKa = 10.4WNGFFITITGEE139 pKa = 3.95LNKK142 pKa = 9.69TKK144 pKa = 10.57SIMEE148 pKa = 3.76GDD150 pKa = 3.71YY151 pKa = 10.59IVRR154 pKa = 11.84CEE156 pKa = 3.89NQLKK160 pKa = 10.17IIHH163 pKa = 6.64KK164 pKa = 9.71LDD166 pKa = 4.42GIIRR170 pKa = 11.84TIQKK174 pKa = 9.94KK175 pKa = 9.73SKK177 pKa = 9.55EE178 pKa = 4.15YY179 pKa = 10.67DD180 pKa = 2.94SSTIKK185 pKa = 10.85NYY187 pKa = 9.84IDD189 pKa = 3.06FTMYY193 pKa = 11.0LDD195 pKa = 3.97DD196 pKa = 4.28CSYY199 pKa = 11.28VVSSSQVRR207 pKa = 11.84RR208 pKa = 11.84NDD210 pKa = 3.09EE211 pKa = 4.16DD212 pKa = 3.76CAEE215 pKa = 4.16TVIDD219 pKa = 4.3EE220 pKa = 4.62NQDD223 pKa = 3.31KK224 pKa = 11.33LNIAEE229 pKa = 4.3SDD231 pKa = 3.48ATT233 pKa = 3.76
Molecular weight: 27.17 kDa
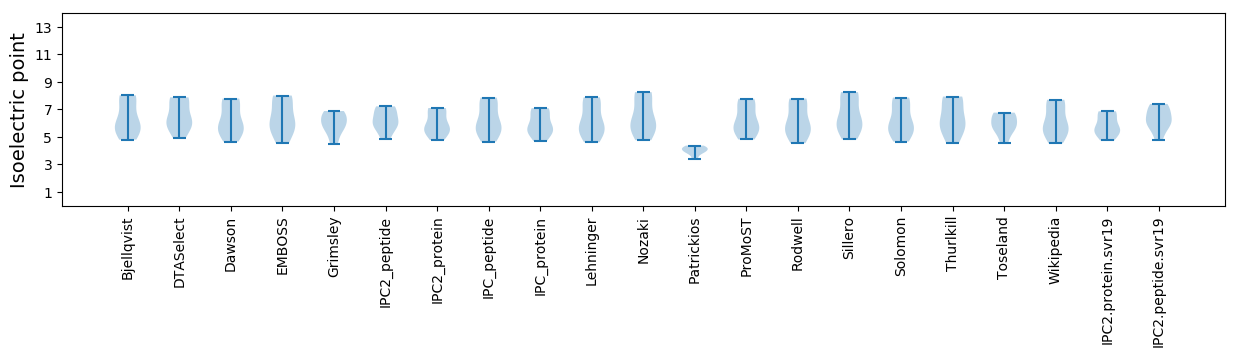
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8K9Y9|F8K9Y9_9VIRU Nucleocapsid OS=Raspberry leaf blotch emaravirus OX=1980431 GN=NC PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 10.48IMVQLILLCILLQFIASEE20 pKa = 4.39SSEE23 pKa = 4.0SRR25 pKa = 11.84PDD27 pKa = 3.51SEE29 pKa = 4.95YY30 pKa = 11.34KK31 pKa = 10.41EE32 pKa = 4.22ADD34 pKa = 3.79EE35 pKa = 5.6NIHH38 pKa = 6.8DD39 pKa = 4.44CSLKK43 pKa = 10.38VVRR46 pKa = 11.84LDD48 pKa = 5.04GIQLACRR55 pKa = 11.84GKK57 pKa = 9.24ILCWFSTNAVAYY69 pKa = 10.81NDD71 pKa = 4.21TCWKK75 pKa = 8.65LTEE78 pKa = 4.32TSTHH82 pKa = 5.35FHH84 pKa = 6.87CCNKK88 pKa = 10.15YY89 pKa = 9.44EE90 pKa = 3.95AVIYY94 pKa = 10.09KK95 pKa = 10.24ISDD98 pKa = 3.18EE99 pKa = 4.35SRR101 pKa = 11.84NICYY105 pKa = 10.3DD106 pKa = 3.44VFSHH110 pKa = 5.78SWCVFLSNLYY120 pKa = 10.49HH121 pKa = 7.11FILAILIWIIMALARR136 pKa = 11.84VPFLYY141 pKa = 10.07IFKK144 pKa = 10.34IIDD147 pKa = 4.57LISGKK152 pKa = 8.99ILKK155 pKa = 9.75KK156 pKa = 10.41KK157 pKa = 10.0KK158 pKa = 10.24SCVKK162 pKa = 10.86CEE164 pKa = 3.65DD165 pKa = 3.41KK166 pKa = 11.31YY167 pKa = 11.65AFFHH171 pKa = 6.82IDD173 pKa = 3.26CSSSTRR179 pKa = 11.84KK180 pKa = 9.6RR181 pKa = 11.84VDD183 pKa = 3.17YY184 pKa = 9.05NTVYY188 pKa = 8.61YY189 pKa = 9.55TIVILVIFLVPVFASGEE206 pKa = 4.05NLKK209 pKa = 11.32DD210 pKa = 3.41NDD212 pKa = 4.53SKK214 pKa = 11.65FNTYY218 pKa = 10.32DD219 pKa = 2.96HH220 pKa = 6.74SGYY223 pKa = 9.38TEE225 pKa = 3.97FVVKK229 pKa = 10.56DD230 pKa = 3.54VEE232 pKa = 4.38HH233 pKa = 6.58EE234 pKa = 4.21VNEE237 pKa = 4.88FYY239 pKa = 10.85TKK241 pKa = 10.13TNHH244 pKa = 5.9IKK246 pKa = 8.7ITIEE250 pKa = 3.55KK251 pKa = 10.08SYY253 pKa = 10.8ISYY256 pKa = 10.25VLSYY260 pKa = 11.02SHH262 pKa = 7.65DD263 pKa = 3.79VLKK266 pKa = 9.78KK267 pKa = 7.76TNDD270 pKa = 3.33VITSLEE276 pKa = 4.03YY277 pKa = 10.97SCDD280 pKa = 3.85SEE282 pKa = 4.5EE283 pKa = 3.72KK284 pKa = 10.33CYY286 pKa = 10.75RR287 pKa = 11.84SLKK290 pKa = 9.63GDD292 pKa = 2.94NRR294 pKa = 11.84TFRR297 pKa = 11.84SLKK300 pKa = 10.0KK301 pKa = 10.01NHH303 pKa = 7.3DD304 pKa = 3.76GLSCIFSNAIVCMACSFHH322 pKa = 7.4YY323 pKa = 10.5EE324 pKa = 4.32LFAEE328 pKa = 4.98VYY330 pKa = 9.28TVTEE334 pKa = 4.29HH335 pKa = 7.18KK336 pKa = 10.36PIILMSVEE344 pKa = 3.94INNSEE349 pKa = 4.41KK350 pKa = 11.22YY351 pKa = 10.72NITLSSYY358 pKa = 11.2NDD360 pKa = 3.46YY361 pKa = 11.16VDD363 pKa = 3.35EE364 pKa = 4.29NFYY367 pKa = 10.63IRR369 pKa = 11.84SLNAVDD375 pKa = 4.21FTDD378 pKa = 6.09DD379 pKa = 2.96IVLIKK384 pKa = 10.89GLVAYY389 pKa = 9.31KK390 pKa = 10.7GKK392 pKa = 9.93ICTVPSLDD400 pKa = 4.65CFGSHH405 pKa = 6.01IKK407 pKa = 10.21KK408 pKa = 10.32DD409 pKa = 3.77NKK411 pKa = 9.49ILAMYY416 pKa = 10.21NPIITDD422 pKa = 3.32NNHH425 pKa = 6.05YY426 pKa = 7.91DD427 pKa = 3.48TTVNLVKK434 pKa = 10.25CTEE437 pKa = 3.98NEE439 pKa = 3.96NLDD442 pKa = 4.05LLRR445 pKa = 11.84LSKK448 pKa = 10.57IGLIVDD454 pKa = 3.66KK455 pKa = 11.1DD456 pKa = 3.62RR457 pKa = 11.84VIEE460 pKa = 4.31DD461 pKa = 3.17RR462 pKa = 11.84SFGYY466 pKa = 10.25FSVGVRR472 pKa = 11.84NKK474 pKa = 9.67MLLDD478 pKa = 3.59NKK480 pKa = 10.04ICEE483 pKa = 3.86KK484 pKa = 10.73DD485 pKa = 3.42AIVRR489 pKa = 11.84SIEE492 pKa = 4.17ANGCYY497 pKa = 9.59NCKK500 pKa = 10.25FGFNVKK506 pKa = 9.73INYY509 pKa = 9.27KK510 pKa = 10.32LYY512 pKa = 10.16SEE514 pKa = 4.69CGKK517 pKa = 9.94IEE519 pKa = 4.56CKK521 pKa = 9.8TPIYY525 pKa = 10.39KK526 pKa = 10.35SYY528 pKa = 10.04TYY530 pKa = 11.03VSNGTTANIKK540 pKa = 10.32LYY542 pKa = 10.88SDD544 pKa = 5.11DD545 pKa = 4.25YY546 pKa = 10.22LTKK549 pKa = 9.29ITCNGKK555 pKa = 9.15EE556 pKa = 3.99VQIKK560 pKa = 10.48LDD562 pKa = 3.51SLNIDD567 pKa = 3.4SYY569 pKa = 9.92YY570 pKa = 10.89HH571 pKa = 6.0NNAYY575 pKa = 6.57TTNNLPIKK583 pKa = 10.54SRR585 pKa = 11.84IKK587 pKa = 10.76NVLNLALYY595 pKa = 10.38DD596 pKa = 3.95SFKK599 pKa = 10.5MAVLTIILILFSYY612 pKa = 9.44TILSILIKK620 pKa = 10.38LRR622 pKa = 11.84RR623 pKa = 11.84LNKK626 pKa = 9.91RR627 pKa = 11.84STHH630 pKa = 5.6KK631 pKa = 11.11VLDD634 pKa = 3.65NTLYY638 pKa = 10.97AVDD641 pKa = 4.04MSHH644 pKa = 7.1LRR646 pKa = 11.84NLGDD650 pKa = 3.28
MM1 pKa = 7.76KK2 pKa = 10.48IMVQLILLCILLQFIASEE20 pKa = 4.39SSEE23 pKa = 4.0SRR25 pKa = 11.84PDD27 pKa = 3.51SEE29 pKa = 4.95YY30 pKa = 11.34KK31 pKa = 10.41EE32 pKa = 4.22ADD34 pKa = 3.79EE35 pKa = 5.6NIHH38 pKa = 6.8DD39 pKa = 4.44CSLKK43 pKa = 10.38VVRR46 pKa = 11.84LDD48 pKa = 5.04GIQLACRR55 pKa = 11.84GKK57 pKa = 9.24ILCWFSTNAVAYY69 pKa = 10.81NDD71 pKa = 4.21TCWKK75 pKa = 8.65LTEE78 pKa = 4.32TSTHH82 pKa = 5.35FHH84 pKa = 6.87CCNKK88 pKa = 10.15YY89 pKa = 9.44EE90 pKa = 3.95AVIYY94 pKa = 10.09KK95 pKa = 10.24ISDD98 pKa = 3.18EE99 pKa = 4.35SRR101 pKa = 11.84NICYY105 pKa = 10.3DD106 pKa = 3.44VFSHH110 pKa = 5.78SWCVFLSNLYY120 pKa = 10.49HH121 pKa = 7.11FILAILIWIIMALARR136 pKa = 11.84VPFLYY141 pKa = 10.07IFKK144 pKa = 10.34IIDD147 pKa = 4.57LISGKK152 pKa = 8.99ILKK155 pKa = 9.75KK156 pKa = 10.41KK157 pKa = 10.0KK158 pKa = 10.24SCVKK162 pKa = 10.86CEE164 pKa = 3.65DD165 pKa = 3.41KK166 pKa = 11.31YY167 pKa = 11.65AFFHH171 pKa = 6.82IDD173 pKa = 3.26CSSSTRR179 pKa = 11.84KK180 pKa = 9.6RR181 pKa = 11.84VDD183 pKa = 3.17YY184 pKa = 9.05NTVYY188 pKa = 8.61YY189 pKa = 9.55TIVILVIFLVPVFASGEE206 pKa = 4.05NLKK209 pKa = 11.32DD210 pKa = 3.41NDD212 pKa = 4.53SKK214 pKa = 11.65FNTYY218 pKa = 10.32DD219 pKa = 2.96HH220 pKa = 6.74SGYY223 pKa = 9.38TEE225 pKa = 3.97FVVKK229 pKa = 10.56DD230 pKa = 3.54VEE232 pKa = 4.38HH233 pKa = 6.58EE234 pKa = 4.21VNEE237 pKa = 4.88FYY239 pKa = 10.85TKK241 pKa = 10.13TNHH244 pKa = 5.9IKK246 pKa = 8.7ITIEE250 pKa = 3.55KK251 pKa = 10.08SYY253 pKa = 10.8ISYY256 pKa = 10.25VLSYY260 pKa = 11.02SHH262 pKa = 7.65DD263 pKa = 3.79VLKK266 pKa = 9.78KK267 pKa = 7.76TNDD270 pKa = 3.33VITSLEE276 pKa = 4.03YY277 pKa = 10.97SCDD280 pKa = 3.85SEE282 pKa = 4.5EE283 pKa = 3.72KK284 pKa = 10.33CYY286 pKa = 10.75RR287 pKa = 11.84SLKK290 pKa = 9.63GDD292 pKa = 2.94NRR294 pKa = 11.84TFRR297 pKa = 11.84SLKK300 pKa = 10.0KK301 pKa = 10.01NHH303 pKa = 7.3DD304 pKa = 3.76GLSCIFSNAIVCMACSFHH322 pKa = 7.4YY323 pKa = 10.5EE324 pKa = 4.32LFAEE328 pKa = 4.98VYY330 pKa = 9.28TVTEE334 pKa = 4.29HH335 pKa = 7.18KK336 pKa = 10.36PIILMSVEE344 pKa = 3.94INNSEE349 pKa = 4.41KK350 pKa = 11.22YY351 pKa = 10.72NITLSSYY358 pKa = 11.2NDD360 pKa = 3.46YY361 pKa = 11.16VDD363 pKa = 3.35EE364 pKa = 4.29NFYY367 pKa = 10.63IRR369 pKa = 11.84SLNAVDD375 pKa = 4.21FTDD378 pKa = 6.09DD379 pKa = 2.96IVLIKK384 pKa = 10.89GLVAYY389 pKa = 9.31KK390 pKa = 10.7GKK392 pKa = 9.93ICTVPSLDD400 pKa = 4.65CFGSHH405 pKa = 6.01IKK407 pKa = 10.21KK408 pKa = 10.32DD409 pKa = 3.77NKK411 pKa = 9.49ILAMYY416 pKa = 10.21NPIITDD422 pKa = 3.32NNHH425 pKa = 6.05YY426 pKa = 7.91DD427 pKa = 3.48TTVNLVKK434 pKa = 10.25CTEE437 pKa = 3.98NEE439 pKa = 3.96NLDD442 pKa = 4.05LLRR445 pKa = 11.84LSKK448 pKa = 10.57IGLIVDD454 pKa = 3.66KK455 pKa = 11.1DD456 pKa = 3.62RR457 pKa = 11.84VIEE460 pKa = 4.31DD461 pKa = 3.17RR462 pKa = 11.84SFGYY466 pKa = 10.25FSVGVRR472 pKa = 11.84NKK474 pKa = 9.67MLLDD478 pKa = 3.59NKK480 pKa = 10.04ICEE483 pKa = 3.86KK484 pKa = 10.73DD485 pKa = 3.42AIVRR489 pKa = 11.84SIEE492 pKa = 4.17ANGCYY497 pKa = 9.59NCKK500 pKa = 10.25FGFNVKK506 pKa = 9.73INYY509 pKa = 9.27KK510 pKa = 10.32LYY512 pKa = 10.16SEE514 pKa = 4.69CGKK517 pKa = 9.94IEE519 pKa = 4.56CKK521 pKa = 9.8TPIYY525 pKa = 10.39KK526 pKa = 10.35SYY528 pKa = 10.04TYY530 pKa = 11.03VSNGTTANIKK540 pKa = 10.32LYY542 pKa = 10.88SDD544 pKa = 5.11DD545 pKa = 4.25YY546 pKa = 10.22LTKK549 pKa = 9.29ITCNGKK555 pKa = 9.15EE556 pKa = 3.99VQIKK560 pKa = 10.48LDD562 pKa = 3.51SLNIDD567 pKa = 3.4SYY569 pKa = 9.92YY570 pKa = 10.89HH571 pKa = 6.0NNAYY575 pKa = 6.57TTNNLPIKK583 pKa = 10.54SRR585 pKa = 11.84IKK587 pKa = 10.76NVLNLALYY595 pKa = 10.38DD596 pKa = 3.95SFKK599 pKa = 10.5MAVLTIILILFSYY612 pKa = 9.44TILSILIKK620 pKa = 10.38LRR622 pKa = 11.84RR623 pKa = 11.84LNKK626 pKa = 9.91RR627 pKa = 11.84STHH630 pKa = 5.6KK631 pKa = 11.11VLDD634 pKa = 3.65NTLYY638 pKa = 10.97AVDD641 pKa = 4.04MSHH644 pKa = 7.1LRR646 pKa = 11.84NLGDD650 pKa = 3.28
Molecular weight: 75.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4693 |
186 |
2295 |
586.6 |
68.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.409 ± 0.52 | 1.811 ± 0.388 |
7.053 ± 0.43 | 6.265 ± 0.312 |
4.581 ± 0.231 | 2.813 ± 0.356 |
1.875 ± 0.198 | 9.205 ± 0.544 |
9.163 ± 0.219 | 9.44 ± 0.385 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.685 ± 0.441 | 8.395 ± 0.373 |
2.365 ± 0.358 | 2.152 ± 0.295 |
3.644 ± 0.278 | 8.161 ± 0.359 |
4.944 ± 0.207 | 5.455 ± 0.654 |
0.533 ± 0.113 | 6.052 ± 0.419 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
