
Mobuck virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; unclassified Orbivirus
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
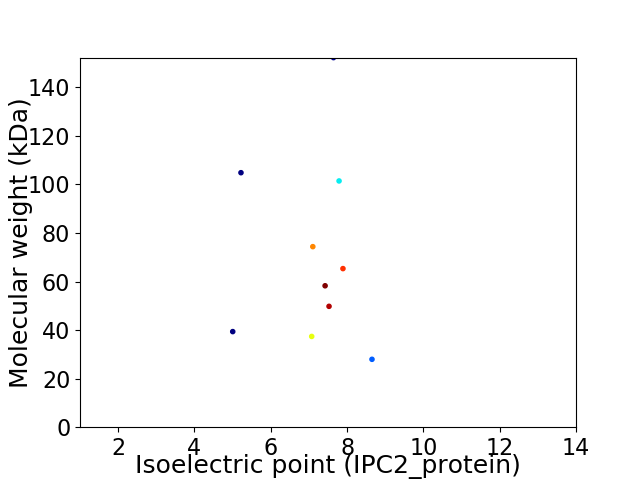
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5NDA7|U5NDA7_9REOV VP3 OS=Mobuck virus OX=1408137 GN=VP3 PE=3 SV=1
MM1 pKa = 7.5EE2 pKa = 6.2GIFSRR7 pKa = 11.84AFCFLEE13 pKa = 3.8CLGANRR19 pKa = 11.84DD20 pKa = 3.26PRR22 pKa = 11.84ARR24 pKa = 11.84RR25 pKa = 11.84TYY27 pKa = 10.52APSDD31 pKa = 3.57GFSLFVTRR39 pKa = 11.84FNATTEE45 pKa = 4.07RR46 pKa = 11.84PITQTPSTVEE56 pKa = 3.5EE57 pKa = 4.31HH58 pKa = 6.49RR59 pKa = 11.84ACFYY63 pKa = 11.34AGLDD67 pKa = 3.67LVIAALGINFNYY79 pKa = 10.32RR80 pKa = 11.84LPNYY84 pKa = 7.84TPNVQVLSILARR96 pKa = 11.84EE97 pKa = 4.13DD98 pKa = 3.51LPYY101 pKa = 8.79TTEE104 pKa = 3.69AFTRR108 pKa = 11.84SSRR111 pKa = 11.84ILNEE115 pKa = 3.73TTAGQSTRR123 pKa = 11.84RR124 pKa = 11.84FTQPWPDD131 pKa = 3.13VSFMYY136 pKa = 10.79LAGTPFRR143 pKa = 11.84TDD145 pKa = 3.36DD146 pKa = 4.51NEE148 pKa = 4.18PQPMAVGYY156 pKa = 11.08NDD158 pKa = 3.65MEE160 pKa = 4.2VTLNVNEE167 pKa = 5.32DD168 pKa = 3.33IEE170 pKa = 4.33ITDD173 pKa = 3.13IVMPNYY179 pKa = 9.51PDD181 pKa = 3.56VVNACITWYY190 pKa = 9.38TLSHH194 pKa = 5.65YY195 pKa = 10.31RR196 pKa = 11.84GAAGALIEE204 pKa = 4.15GSRR207 pKa = 11.84LCTLMIGDD215 pKa = 4.55EE216 pKa = 4.14EE217 pKa = 4.36VDD219 pKa = 3.39AGVEE223 pKa = 4.26LIVPSGTAIRR233 pKa = 11.84VANNSGVNTGIIHH246 pKa = 6.51ITLKK250 pKa = 9.84WFTRR254 pKa = 11.84AQPGEE259 pKa = 4.1HH260 pKa = 6.91LYY262 pKa = 9.68DD263 pKa = 3.62TMLMDD268 pKa = 5.45IDD270 pKa = 3.89ACYY273 pKa = 10.66SFHH276 pKa = 8.85SEE278 pKa = 3.14DD279 pKa = 3.51WYY281 pKa = 10.47EE282 pKa = 3.89LRR284 pKa = 11.84SFLLRR289 pKa = 11.84SVGLPTHH296 pKa = 7.15APPLEE301 pKa = 4.77PIRR304 pKa = 11.84VPNRR308 pKa = 11.84VLALALIARR317 pKa = 11.84LQNVYY322 pKa = 9.7VAHH325 pKa = 6.38SPIDD329 pKa = 3.11IRR331 pKa = 11.84AIQDD335 pKa = 3.17VNGQGGLPNAALEE348 pKa = 4.07ALRR351 pKa = 11.84RR352 pKa = 11.84VV353 pKa = 3.65
MM1 pKa = 7.5EE2 pKa = 6.2GIFSRR7 pKa = 11.84AFCFLEE13 pKa = 3.8CLGANRR19 pKa = 11.84DD20 pKa = 3.26PRR22 pKa = 11.84ARR24 pKa = 11.84RR25 pKa = 11.84TYY27 pKa = 10.52APSDD31 pKa = 3.57GFSLFVTRR39 pKa = 11.84FNATTEE45 pKa = 4.07RR46 pKa = 11.84PITQTPSTVEE56 pKa = 3.5EE57 pKa = 4.31HH58 pKa = 6.49RR59 pKa = 11.84ACFYY63 pKa = 11.34AGLDD67 pKa = 3.67LVIAALGINFNYY79 pKa = 10.32RR80 pKa = 11.84LPNYY84 pKa = 7.84TPNVQVLSILARR96 pKa = 11.84EE97 pKa = 4.13DD98 pKa = 3.51LPYY101 pKa = 8.79TTEE104 pKa = 3.69AFTRR108 pKa = 11.84SSRR111 pKa = 11.84ILNEE115 pKa = 3.73TTAGQSTRR123 pKa = 11.84RR124 pKa = 11.84FTQPWPDD131 pKa = 3.13VSFMYY136 pKa = 10.79LAGTPFRR143 pKa = 11.84TDD145 pKa = 3.36DD146 pKa = 4.51NEE148 pKa = 4.18PQPMAVGYY156 pKa = 11.08NDD158 pKa = 3.65MEE160 pKa = 4.2VTLNVNEE167 pKa = 5.32DD168 pKa = 3.33IEE170 pKa = 4.33ITDD173 pKa = 3.13IVMPNYY179 pKa = 9.51PDD181 pKa = 3.56VVNACITWYY190 pKa = 9.38TLSHH194 pKa = 5.65YY195 pKa = 10.31RR196 pKa = 11.84GAAGALIEE204 pKa = 4.15GSRR207 pKa = 11.84LCTLMIGDD215 pKa = 4.55EE216 pKa = 4.14EE217 pKa = 4.36VDD219 pKa = 3.39AGVEE223 pKa = 4.26LIVPSGTAIRR233 pKa = 11.84VANNSGVNTGIIHH246 pKa = 6.51ITLKK250 pKa = 9.84WFTRR254 pKa = 11.84AQPGEE259 pKa = 4.1HH260 pKa = 6.91LYY262 pKa = 9.68DD263 pKa = 3.62TMLMDD268 pKa = 5.45IDD270 pKa = 3.89ACYY273 pKa = 10.66SFHH276 pKa = 8.85SEE278 pKa = 3.14DD279 pKa = 3.51WYY281 pKa = 10.47EE282 pKa = 3.89LRR284 pKa = 11.84SFLLRR289 pKa = 11.84SVGLPTHH296 pKa = 7.15APPLEE301 pKa = 4.77PIRR304 pKa = 11.84VPNRR308 pKa = 11.84VLALALIARR317 pKa = 11.84LQNVYY322 pKa = 9.7VAHH325 pKa = 6.38SPIDD329 pKa = 3.11IRR331 pKa = 11.84AIQDD335 pKa = 3.17VNGQGGLPNAALEE348 pKa = 4.07ALRR351 pKa = 11.84RR352 pKa = 11.84VV353 pKa = 3.65
Molecular weight: 39.38 kDa
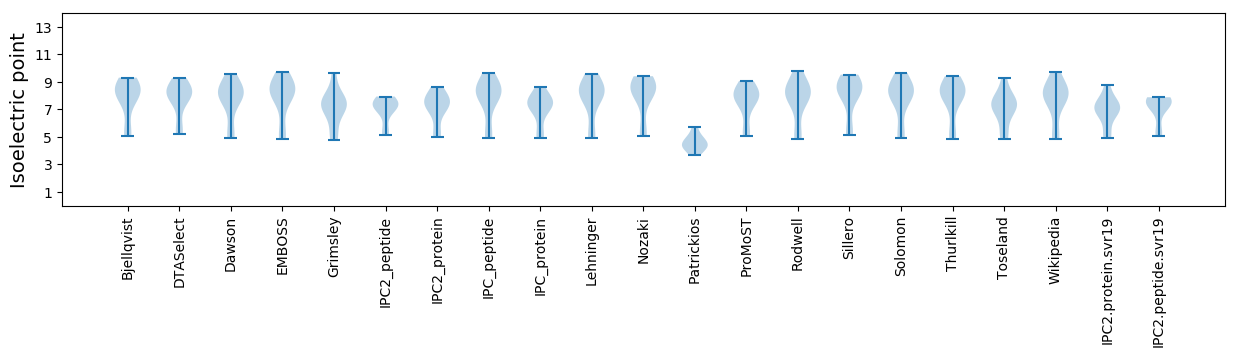
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5NGR3|U5NGR3_9REOV Non-structural protein NS3 OS=Mobuck virus OX=1408137 GN=Nsp3 PE=3 SV=1
MM1 pKa = 7.33HH2 pKa = 7.65AAIQPVKK9 pKa = 10.36RR10 pKa = 11.84EE11 pKa = 3.97NPIPLLTVVPASAPAYY27 pKa = 8.11EE28 pKa = 4.42KK29 pKa = 10.98VFHH32 pKa = 7.08EE33 pKa = 4.25EE34 pKa = 3.96TKK36 pKa = 10.83QKK38 pKa = 8.79EE39 pKa = 4.3TLLNLTVPDD48 pKa = 4.13ALTARR53 pKa = 11.84TTALDD58 pKa = 3.63VLSNALGAGSGTDD71 pKa = 3.0EE72 pKa = 3.66ITRR75 pKa = 11.84RR76 pKa = 11.84EE77 pKa = 3.76RR78 pKa = 11.84SAYY81 pKa = 9.36GAAAQALNEE90 pKa = 4.4DD91 pKa = 3.88EE92 pKa = 4.31STRR95 pKa = 11.84RR96 pKa = 11.84LKK98 pKa = 11.04VYY100 pKa = 9.07MNRR103 pKa = 11.84QIIPRR108 pKa = 11.84LEE110 pKa = 3.82QKK112 pKa = 10.64YY113 pKa = 7.88RR114 pKa = 11.84TSTIKK119 pKa = 10.34YY120 pKa = 8.26RR121 pKa = 11.84LWVCVEE127 pKa = 3.61IFFAFLSIMIFGVMGVTEE145 pKa = 4.18WHH147 pKa = 6.53EE148 pKa = 5.44SIDD151 pKa = 3.79DD152 pKa = 3.65WMKK155 pKa = 9.68KK156 pKa = 7.87QSGIKK161 pKa = 10.17VGVSTVSMICTMILLYY177 pKa = 10.58ASRR180 pKa = 11.84SAGGLKK186 pKa = 9.77EE187 pKa = 3.64LRR189 pKa = 11.84KK190 pKa = 9.42RR191 pKa = 11.84LKK193 pKa = 10.32RR194 pKa = 11.84EE195 pKa = 3.74LIKK198 pKa = 10.8RR199 pKa = 11.84QTYY202 pKa = 10.38NDD204 pKa = 2.99ISGTMGGTEE213 pKa = 3.99THH215 pKa = 6.34GGEE218 pKa = 4.08SDD220 pKa = 3.36SRR222 pKa = 11.84LSNRR226 pKa = 11.84EE227 pKa = 3.01NSAWINAPGVSSPASGWQLMRR248 pKa = 11.84VDD250 pKa = 4.03GG251 pKa = 4.37
MM1 pKa = 7.33HH2 pKa = 7.65AAIQPVKK9 pKa = 10.36RR10 pKa = 11.84EE11 pKa = 3.97NPIPLLTVVPASAPAYY27 pKa = 8.11EE28 pKa = 4.42KK29 pKa = 10.98VFHH32 pKa = 7.08EE33 pKa = 4.25EE34 pKa = 3.96TKK36 pKa = 10.83QKK38 pKa = 8.79EE39 pKa = 4.3TLLNLTVPDD48 pKa = 4.13ALTARR53 pKa = 11.84TTALDD58 pKa = 3.63VLSNALGAGSGTDD71 pKa = 3.0EE72 pKa = 3.66ITRR75 pKa = 11.84RR76 pKa = 11.84EE77 pKa = 3.76RR78 pKa = 11.84SAYY81 pKa = 9.36GAAAQALNEE90 pKa = 4.4DD91 pKa = 3.88EE92 pKa = 4.31STRR95 pKa = 11.84RR96 pKa = 11.84LKK98 pKa = 11.04VYY100 pKa = 9.07MNRR103 pKa = 11.84QIIPRR108 pKa = 11.84LEE110 pKa = 3.82QKK112 pKa = 10.64YY113 pKa = 7.88RR114 pKa = 11.84TSTIKK119 pKa = 10.34YY120 pKa = 8.26RR121 pKa = 11.84LWVCVEE127 pKa = 3.61IFFAFLSIMIFGVMGVTEE145 pKa = 4.18WHH147 pKa = 6.53EE148 pKa = 5.44SIDD151 pKa = 3.79DD152 pKa = 3.65WMKK155 pKa = 9.68KK156 pKa = 7.87QSGIKK161 pKa = 10.17VGVSTVSMICTMILLYY177 pKa = 10.58ASRR180 pKa = 11.84SAGGLKK186 pKa = 9.77EE187 pKa = 3.64LRR189 pKa = 11.84KK190 pKa = 9.42RR191 pKa = 11.84LKK193 pKa = 10.32RR194 pKa = 11.84EE195 pKa = 3.74LIKK198 pKa = 10.8RR199 pKa = 11.84QTYY202 pKa = 10.38NDD204 pKa = 2.99ISGTMGGTEE213 pKa = 3.99THH215 pKa = 6.34GGEE218 pKa = 4.08SDD220 pKa = 3.36SRR222 pKa = 11.84LSNRR226 pKa = 11.84EE227 pKa = 3.01NSAWINAPGVSSPASGWQLMRR248 pKa = 11.84VDD250 pKa = 4.03GG251 pKa = 4.37
Molecular weight: 27.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6255 |
251 |
1328 |
625.5 |
71.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.347 ± 0.564 | 1.503 ± 0.184 |
5.675 ± 0.43 | 6.938 ± 0.48 |
3.949 ± 0.365 | 5.851 ± 0.237 |
2.126 ± 0.187 | 6.843 ± 0.275 |
6.027 ± 0.743 | 9.081 ± 0.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.469 ± 0.132 | 4.317 ± 0.314 |
3.837 ± 0.446 | 3.357 ± 0.286 |
6.763 ± 0.241 | 6.491 ± 0.388 |
6.347 ± 0.357 | 6.507 ± 0.273 |
1.055 ± 0.19 | 3.517 ± 0.344 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
