
Chitinophaga pinensis (strain ATCC 43595 / DSM 2588 / LMG 13176 / NBRC 15968 / NCIMB 11800 / UQM 2034)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Chitinophaga; Chitinophaga pinensis
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
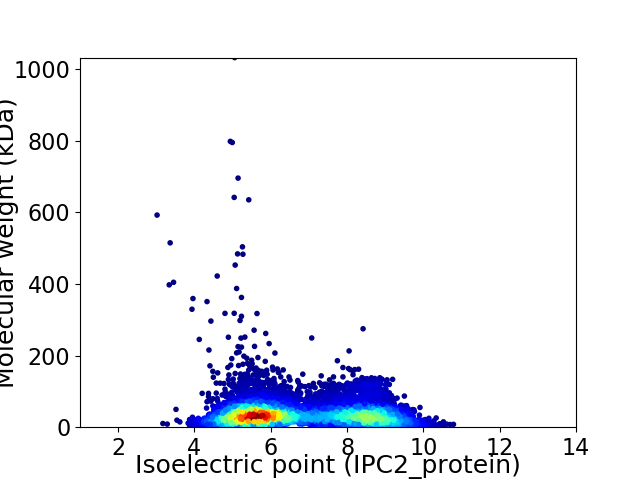
Virtual 2D-PAGE plot for 7179 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7PSK9|C7PSK9_CHIPD Lipid-A-disaccharide synthase OS=Chitinophaga pinensis (strain ATCC 43595 / DSM 2588 / LMG 13176 / NBRC 15968 / NCIMB 11800 / UQM 2034) OX=485918 GN=Cpin_7057 PE=4 SV=1
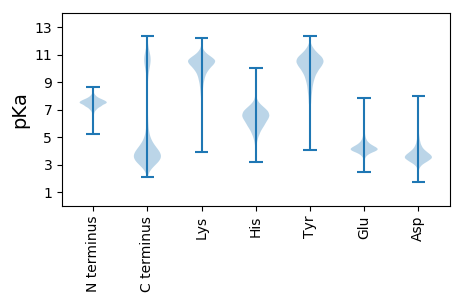
MM1 pKa = 7.69EE2 pKa = 5.08EE3 pKa = 4.33FYY5 pKa = 11.22DD6 pKa = 4.49AASEE10 pKa = 4.38KK11 pKa = 9.76YY12 pKa = 9.79KK13 pKa = 10.75KK14 pKa = 10.57YY15 pKa = 10.61LLQVVYY21 pKa = 10.84NDD23 pKa = 3.13EE24 pKa = 4.05TYY26 pKa = 9.25YY27 pKa = 10.36TVSGADD33 pKa = 3.34LSDD36 pKa = 3.96NEE38 pKa = 4.28ATRR41 pKa = 11.84LLTDD45 pKa = 3.94ADD47 pKa = 4.21GKK49 pKa = 10.17ICLYY53 pKa = 11.13ADD55 pKa = 4.13LPSLRR60 pKa = 11.84KK61 pKa = 10.02GIEE64 pKa = 3.86AGVVTFDD71 pKa = 4.71TPNLQAWGKK80 pKa = 10.11DD81 pKa = 3.03INEE84 pKa = 4.09TDD86 pKa = 3.22TAYY89 pKa = 9.91TGVDD93 pKa = 3.99FFSLKK98 pKa = 10.28SEE100 pKa = 4.14HH101 pKa = 7.03LEE103 pKa = 4.45ADD105 pKa = 4.32DD106 pKa = 6.32DD107 pKa = 4.37PLLYY111 pKa = 10.31EE112 pKa = 4.46IYY114 pKa = 10.45GALGVVRR121 pKa = 11.84DD122 pKa = 3.84YY123 pKa = 11.89AEE125 pKa = 3.95QEE127 pKa = 4.08NATEE131 pKa = 5.0LLTLLDD137 pKa = 3.88SPIVNEE143 pKa = 3.89YY144 pKa = 10.26MEE146 pKa = 4.3ICADD150 pKa = 4.05LFLWSSDD157 pKa = 2.78TDD159 pKa = 3.73SFRR162 pKa = 11.84EE163 pKa = 4.02DD164 pKa = 3.01FDD166 pKa = 5.35FNAFVPVLGQIYY178 pKa = 8.4TLLEE182 pKa = 3.66PRR184 pKa = 11.84LRR186 pKa = 11.84VVV188 pKa = 3.33
MM1 pKa = 7.69EE2 pKa = 5.08EE3 pKa = 4.33FYY5 pKa = 11.22DD6 pKa = 4.49AASEE10 pKa = 4.38KK11 pKa = 9.76YY12 pKa = 9.79KK13 pKa = 10.75KK14 pKa = 10.57YY15 pKa = 10.61LLQVVYY21 pKa = 10.84NDD23 pKa = 3.13EE24 pKa = 4.05TYY26 pKa = 9.25YY27 pKa = 10.36TVSGADD33 pKa = 3.34LSDD36 pKa = 3.96NEE38 pKa = 4.28ATRR41 pKa = 11.84LLTDD45 pKa = 3.94ADD47 pKa = 4.21GKK49 pKa = 10.17ICLYY53 pKa = 11.13ADD55 pKa = 4.13LPSLRR60 pKa = 11.84KK61 pKa = 10.02GIEE64 pKa = 3.86AGVVTFDD71 pKa = 4.71TPNLQAWGKK80 pKa = 10.11DD81 pKa = 3.03INEE84 pKa = 4.09TDD86 pKa = 3.22TAYY89 pKa = 9.91TGVDD93 pKa = 3.99FFSLKK98 pKa = 10.28SEE100 pKa = 4.14HH101 pKa = 7.03LEE103 pKa = 4.45ADD105 pKa = 4.32DD106 pKa = 6.32DD107 pKa = 4.37PLLYY111 pKa = 10.31EE112 pKa = 4.46IYY114 pKa = 10.45GALGVVRR121 pKa = 11.84DD122 pKa = 3.84YY123 pKa = 11.89AEE125 pKa = 3.95QEE127 pKa = 4.08NATEE131 pKa = 5.0LLTLLDD137 pKa = 3.88SPIVNEE143 pKa = 3.89YY144 pKa = 10.26MEE146 pKa = 4.3ICADD150 pKa = 4.05LFLWSSDD157 pKa = 2.78TDD159 pKa = 3.73SFRR162 pKa = 11.84EE163 pKa = 4.02DD164 pKa = 3.01FDD166 pKa = 5.35FNAFVPVLGQIYY178 pKa = 8.4TLLEE182 pKa = 3.66PRR184 pKa = 11.84LRR186 pKa = 11.84VVV188 pKa = 3.33
Molecular weight: 21.34 kDa
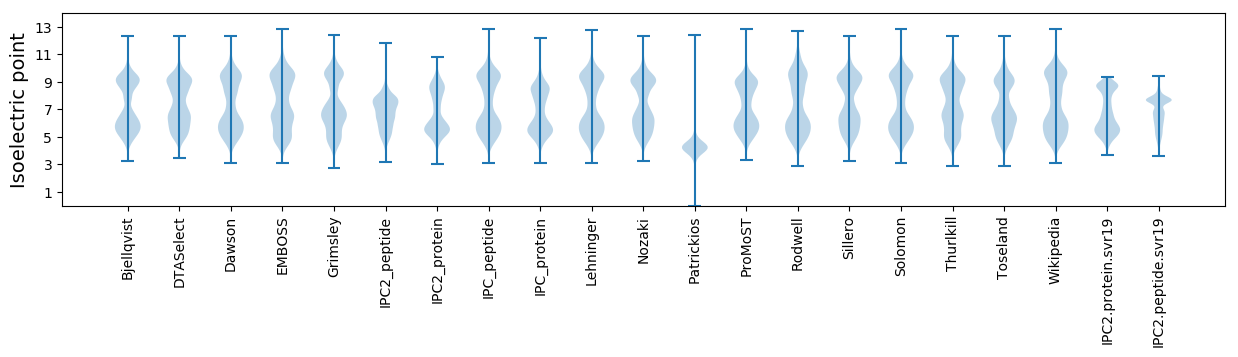
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7PEB3|C7PEB3_CHIPD Uncharacterized protein OS=Chitinophaga pinensis (strain ATCC 43595 / DSM 2588 / LMG 13176 / NBRC 15968 / NCIMB 11800 / UQM 2034) OX=485918 GN=Cpin_5904 PE=4 SV=1
MM1 pKa = 7.04SRR3 pKa = 11.84EE4 pKa = 3.96YY5 pKa = 10.17FKK7 pKa = 11.21SIIRR11 pKa = 11.84TLRR14 pKa = 11.84VSFQVLQNNDD24 pKa = 3.75PLRR27 pKa = 11.84LAGATAFFASFALPPILLILLQVFGLLFDD56 pKa = 3.8RR57 pKa = 11.84RR58 pKa = 11.84SIGRR62 pKa = 11.84QLIGRR67 pKa = 11.84LGEE70 pKa = 4.2LVGKK74 pKa = 10.29DD75 pKa = 2.87SAQQVVITLRR85 pKa = 11.84GFRR88 pKa = 11.84GLAQTWPIAIILFVFLLFVATTLLKK113 pKa = 10.65VIRR116 pKa = 11.84SSLNQVWMIRR126 pKa = 11.84IAVRR130 pKa = 11.84EE131 pKa = 4.11SFLDD135 pKa = 3.48SLLGRR140 pKa = 11.84LRR142 pKa = 11.84SLIVIGAAGLLFLASVLGEE161 pKa = 4.13SAHH164 pKa = 6.74AFLGNYY170 pKa = 8.93VNEE173 pKa = 4.79LFPSAAFIFSGVFTQLVSLLIVTCWFAVLFRR204 pKa = 11.84YY205 pKa = 10.09LPDD208 pKa = 4.5ARR210 pKa = 11.84PTWKK214 pKa = 10.44VAFSGALLTGVLFSIGKK231 pKa = 9.16LVLRR235 pKa = 11.84RR236 pKa = 11.84LLYY239 pKa = 10.55GGNIGILYY247 pKa = 8.77GASASAVLLMLFIFYY262 pKa = 10.51SAIIFYY268 pKa = 10.71YY269 pKa = 9.89GAAFTKK275 pKa = 9.16VWSEE279 pKa = 4.03FKK281 pKa = 10.35HH282 pKa = 7.39RR283 pKa = 11.84PMQPLRR289 pKa = 11.84HH290 pKa = 4.99ATFYY294 pKa = 11.14EE295 pKa = 4.13LADD298 pKa = 3.51VEE300 pKa = 4.43ARR302 pKa = 3.61
MM1 pKa = 7.04SRR3 pKa = 11.84EE4 pKa = 3.96YY5 pKa = 10.17FKK7 pKa = 11.21SIIRR11 pKa = 11.84TLRR14 pKa = 11.84VSFQVLQNNDD24 pKa = 3.75PLRR27 pKa = 11.84LAGATAFFASFALPPILLILLQVFGLLFDD56 pKa = 3.8RR57 pKa = 11.84RR58 pKa = 11.84SIGRR62 pKa = 11.84QLIGRR67 pKa = 11.84LGEE70 pKa = 4.2LVGKK74 pKa = 10.29DD75 pKa = 2.87SAQQVVITLRR85 pKa = 11.84GFRR88 pKa = 11.84GLAQTWPIAIILFVFLLFVATTLLKK113 pKa = 10.65VIRR116 pKa = 11.84SSLNQVWMIRR126 pKa = 11.84IAVRR130 pKa = 11.84EE131 pKa = 4.11SFLDD135 pKa = 3.48SLLGRR140 pKa = 11.84LRR142 pKa = 11.84SLIVIGAAGLLFLASVLGEE161 pKa = 4.13SAHH164 pKa = 6.74AFLGNYY170 pKa = 8.93VNEE173 pKa = 4.79LFPSAAFIFSGVFTQLVSLLIVTCWFAVLFRR204 pKa = 11.84YY205 pKa = 10.09LPDD208 pKa = 4.5ARR210 pKa = 11.84PTWKK214 pKa = 10.44VAFSGALLTGVLFSIGKK231 pKa = 9.16LVLRR235 pKa = 11.84RR236 pKa = 11.84LLYY239 pKa = 10.55GGNIGILYY247 pKa = 8.77GASASAVLLMLFIFYY262 pKa = 10.51SAIIFYY268 pKa = 10.71YY269 pKa = 9.89GAAFTKK275 pKa = 9.16VWSEE279 pKa = 4.03FKK281 pKa = 10.35HH282 pKa = 7.39RR283 pKa = 11.84PMQPLRR289 pKa = 11.84HH290 pKa = 4.99ATFYY294 pKa = 11.14EE295 pKa = 4.13LADD298 pKa = 3.51VEE300 pKa = 4.43ARR302 pKa = 3.61
Molecular weight: 33.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2679255 |
30 |
9175 |
373.2 |
41.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.109 ± 0.036 | 0.837 ± 0.012 |
5.364 ± 0.023 | 5.477 ± 0.036 |
4.524 ± 0.025 | 6.966 ± 0.03 |
2.039 ± 0.019 | 6.611 ± 0.026 |
5.951 ± 0.037 | 9.518 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.018 | 5.113 ± 0.041 |
4.121 ± 0.018 | 4.091 ± 0.021 |
4.475 ± 0.025 | 6.201 ± 0.028 |
6.188 ± 0.059 | 6.533 ± 0.032 |
1.282 ± 0.013 | 4.228 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
