
Bradyrhizobium erythrophlei
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; Bradyrhizobium
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
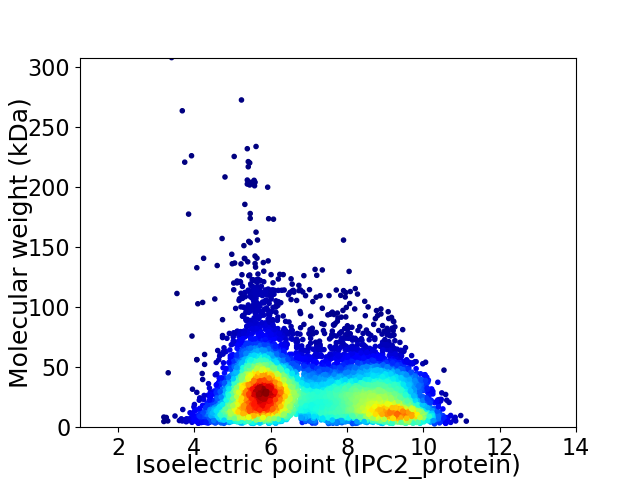
Virtual 2D-PAGE plot for 10605 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N6G2X9|A0A1N6G2X9_9BRAD Carbon-monoxide dehydrogenase medium subunit OS=Bradyrhizobium erythrophlei OX=1437360 GN=SAMN05443247_01429 PE=4 SV=1
MM1 pKa = 6.89STIPAFTFGGITRR14 pKa = 11.84PALILDD20 pKa = 3.7PSGNIILDD28 pKa = 3.51PAAQAFLDD36 pKa = 4.13TYY38 pKa = 8.75GTKK41 pKa = 10.45ALYY44 pKa = 10.61AGVPPGTHH52 pKa = 6.69FPPIPDD58 pKa = 4.32LLTTPVDD65 pKa = 3.88TNGTANSVAEE75 pKa = 4.3GAAVNTLVGITASSHH90 pKa = 5.49SLLPFSITYY99 pKa = 10.27SLSADD104 pKa = 3.43SSSGGFKK111 pKa = 9.66IDD113 pKa = 3.28PTTGVVSVANSAKK126 pKa = 9.68IDD128 pKa = 3.82FEE130 pKa = 4.5SSPGHH135 pKa = 6.85AYY137 pKa = 8.76TVTVQATDD145 pKa = 4.6GIFTTSQNFTIGVSDD160 pKa = 3.95VAPSTPVDD168 pKa = 3.49GNAATNTIAEE178 pKa = 4.44GAAVGTAVGVTAHH191 pKa = 6.0STDD194 pKa = 3.34INGGTVTYY202 pKa = 10.6SLTGDD207 pKa = 3.4TSGGGFTVNAATGVVTVADD226 pKa = 3.81STKK229 pKa = 9.92IDD231 pKa = 3.79YY232 pKa = 7.64EE233 pKa = 4.35TAPGHH238 pKa = 6.26AYY240 pKa = 10.39SVTVQASDD248 pKa = 3.46GTLTSATQTFTIAVSDD264 pKa = 4.15VPLPTPADD272 pKa = 3.69TNPAANQVAEE282 pKa = 4.45GAAAGTLVGLTATANDD298 pKa = 4.21PNGPTTTYY306 pKa = 11.34SLIGDD311 pKa = 4.04TSGGGFTIDD320 pKa = 3.51PATGVVTVADD330 pKa = 3.69PAKK333 pKa = 10.28IDD335 pKa = 3.59FEE337 pKa = 4.64SAAGHH342 pKa = 6.6AYY344 pKa = 10.33SVTVKK349 pKa = 10.79ADD351 pKa = 3.7DD352 pKa = 4.45GLASTTQTFSIAVTDD367 pKa = 3.99VAPSAPTDD375 pKa = 3.45SDD377 pKa = 3.62ASANTIAEE385 pKa = 4.28GAANGSTVGITAHH398 pKa = 5.68STDD401 pKa = 3.41VNGPAVTYY409 pKa = 10.67SLVGDD414 pKa = 3.99TSGGGFTINAATGVITVADD433 pKa = 3.82STKK436 pKa = 9.89IDD438 pKa = 3.83YY439 pKa = 7.68EE440 pKa = 4.29TSPGHH445 pKa = 6.9AYY447 pKa = 8.8TVTAQASDD455 pKa = 3.54GTLASSQTFTIGVTDD470 pKa = 4.06VPPSNPVDD478 pKa = 3.84SNAAANAVAEE488 pKa = 4.54GAAAGATVGITASSTDD504 pKa = 3.35VNGPAVTYY512 pKa = 10.68SLTGDD517 pKa = 3.4TSGGGFTINSATGVVTVADD536 pKa = 3.88PTKK539 pKa = 10.0IDD541 pKa = 3.74YY542 pKa = 9.46EE543 pKa = 4.44SSPGHH548 pKa = 6.67AYY550 pKa = 10.64SITVQSSDD558 pKa = 3.08GTLTSSQVFTIGVNDD573 pKa = 3.8VAPSTPVDD581 pKa = 3.52SDD583 pKa = 3.19ATANAVAEE591 pKa = 4.49GAAAGTNVGITAHH604 pKa = 6.25SVDD607 pKa = 3.78VNGPAVTYY615 pKa = 10.67SLVGDD620 pKa = 3.99TSGGGFTINSATGVVTVADD639 pKa = 3.56PTKK642 pKa = 10.37IDD644 pKa = 3.84FEE646 pKa = 4.89SSGGSYY652 pKa = 9.14TVVAQSTDD660 pKa = 3.12GTLTNSQAFVIAVTDD675 pKa = 3.77VAPSTPVDD683 pKa = 3.74SNAAANTIAEE693 pKa = 4.36GAAAGSTVGITASSTDD709 pKa = 3.35VNGPAVTYY717 pKa = 10.67SLVGDD722 pKa = 3.99TSGGGFTINATTGVVTVTDD741 pKa = 3.77PTKK744 pKa = 10.13IDD746 pKa = 3.54YY747 pKa = 10.42EE748 pKa = 4.44SAPGHH753 pKa = 6.79AYY755 pKa = 8.84TVTSQASDD763 pKa = 3.4GTLTSSQAFTIAVTDD778 pKa = 3.6VAPSVPVDD786 pKa = 3.31SDD788 pKa = 3.42AAANAVTEE796 pKa = 4.16GAPAGSTVGVTASSTDD812 pKa = 3.38VNGPPVTYY820 pKa = 10.49SLTGDD825 pKa = 3.4TSGGGFTINATTGVVTVADD844 pKa = 4.16PSKK847 pKa = 10.46IDD849 pKa = 3.47YY850 pKa = 10.22EE851 pKa = 4.48SAPGHH856 pKa = 6.4AYY858 pKa = 10.34SVTVTSSDD866 pKa = 2.82GTMANSQVFTIAVNDD881 pKa = 4.12AAPSTPVDD889 pKa = 3.27SDD891 pKa = 3.44AAANTVVEE899 pKa = 4.72GAAAGTTVGVTASSTDD915 pKa = 3.33VNGPAVTYY923 pKa = 10.68SLTGDD928 pKa = 3.4TSGGGFTINAATGVVTVADD947 pKa = 3.56PTKK950 pKa = 10.4IDD952 pKa = 4.04FEE954 pKa = 4.72STAPGHH960 pKa = 6.27TYY962 pKa = 9.93TVTATASDD970 pKa = 3.79GTLASSQIFTIAVTDD985 pKa = 4.47APPSTPVDD993 pKa = 3.16SDD995 pKa = 3.69GAINQVSQNAPVGTYY1010 pKa = 9.26TGLTASSTDD1019 pKa = 3.33VNGPAVTYY1027 pKa = 10.67SLVGDD1032 pKa = 3.97TSGGGFAIDD1041 pKa = 3.44ASTGKK1046 pKa = 8.5VTVADD1051 pKa = 3.61STKK1054 pKa = 9.96IPFNAGSPSFDD1065 pKa = 3.15VTVQSSDD1072 pKa = 3.39GTDD1075 pKa = 2.98TSQQTFTINVIQNTPPVANDD1095 pKa = 3.78DD1096 pKa = 4.05SLSATEE1102 pKa = 5.61AGGLNNSIPGSNPSGNVILGTGSAGAVQDD1131 pKa = 4.3TDD1133 pKa = 3.73AQDD1136 pKa = 3.5PSTALTVVAVHH1147 pKa = 6.6TGPEE1151 pKa = 4.29GGSTGTGTVGSPIVGLHH1168 pKa = 5.31GTLTLDD1174 pKa = 3.06SHH1176 pKa = 6.64GAYY1179 pKa = 9.26TYY1181 pKa = 9.67TVNQTDD1187 pKa = 3.69PTVQALHH1194 pKa = 6.12TAAEE1198 pKa = 4.5TITDD1202 pKa = 3.56QFNYY1206 pKa = 9.51TIQDD1210 pKa = 3.32TGGLQDD1216 pKa = 3.34TATITFTIHH1225 pKa = 6.7GADD1228 pKa = 5.0DD1229 pKa = 4.41LPQAIADD1236 pKa = 3.96TGTMTEE1242 pKa = 4.78DD1243 pKa = 4.38DD1244 pKa = 4.91APTLFNVLANDD1255 pKa = 4.09TLDD1258 pKa = 4.45PDD1260 pKa = 3.68HH1261 pKa = 6.8TALNTIAVGPGSITVTGPAGEE1282 pKa = 4.5TFANTDD1288 pKa = 2.74ATATVVSNEE1297 pKa = 4.09VQVSLNNAHH1306 pKa = 6.32FQQLAVGEE1314 pKa = 4.55HH1315 pKa = 5.31ATITVPYY1322 pKa = 9.14TLTGDD1327 pKa = 3.43TGEE1330 pKa = 4.22TSTANLVVTVNGVNDD1345 pKa = 3.47VPVAVDD1351 pKa = 3.86DD1352 pKa = 4.35SGSMTEE1358 pKa = 4.51DD1359 pKa = 3.27QSGHH1363 pKa = 6.24AFTVLSNDD1371 pKa = 3.74TLDD1374 pKa = 4.77PDD1376 pKa = 3.82HH1377 pKa = 6.9GAPNNVTAGAVTNLSAPAGEE1397 pKa = 5.37GITTADD1403 pKa = 2.8IGVSVNASNQVVVNLGANFQHH1424 pKa = 6.09MQDD1427 pKa = 3.67GQTATFDD1434 pKa = 3.48VAYY1437 pKa = 8.76TLHH1440 pKa = 7.18GDD1442 pKa = 3.7QPGDD1446 pKa = 3.12TSTAEE1451 pKa = 3.77LHH1453 pKa = 5.15VTVTGVNDD1461 pKa = 3.53APVAGNFTFNGVNSATGNTDD1481 pKa = 5.28LIVNDD1486 pKa = 4.46PATTAITEE1494 pKa = 4.24TTPNKK1499 pKa = 9.61TISASLLSGASDD1511 pKa = 3.43VDD1513 pKa = 3.92GPGPLTVVAGTIATAHH1529 pKa = 6.31GGSVTMQSDD1538 pKa = 3.47GDD1540 pKa = 4.34FIYY1543 pKa = 10.36KK1544 pKa = 10.23PPVGYY1549 pKa = 8.84TGSDD1553 pKa = 3.13SFNYY1557 pKa = 9.32QVSDD1561 pKa = 3.6QNTGASGVGLGTGTVTINVAAPKK1584 pKa = 9.48VWYY1587 pKa = 10.41VNADD1591 pKa = 3.18AATDD1595 pKa = 3.83GDD1597 pKa = 4.39GSSEE1601 pKa = 4.09HH1602 pKa = 6.91PFNNLTHH1609 pKa = 6.67FNGAGGVDD1617 pKa = 3.68GAGDD1621 pKa = 4.07TIVLEE1626 pKa = 4.48TATAHH1631 pKa = 4.53YY1632 pKa = 9.47TGGLTLEE1639 pKa = 4.48NNEE1642 pKa = 3.97QLISQSAGVTINGTTLFAASGANAVVDD1669 pKa = 3.87GGVVLGSGNTIEE1681 pKa = 5.24GVDD1684 pKa = 4.19FGTTSGFAVSGSSVGTLHH1702 pKa = 7.37LNDD1705 pKa = 3.32THH1707 pKa = 6.65TGVINNSSGGGISIGGASNVLNVDD1731 pKa = 4.61FNSITTGGGTNGIALTNSSGTFHH1754 pKa = 7.02AHH1756 pKa = 6.79GGTISNASGADD1767 pKa = 3.29VALNGGTVTFTDD1779 pKa = 4.46DD1780 pKa = 3.69GAISDD1785 pKa = 3.86STGTVVSVANMTGNTQAFTGAIGTTGSTDD1814 pKa = 3.1GAIALSSNTGATINFSGGMALSTGASDD1841 pKa = 4.31AFSATGGGTVNVTGTNHH1858 pKa = 6.39LTTTTGTALNISNTTIGSSNVTFHH1882 pKa = 7.67DD1883 pKa = 3.37ISSNGGSSNGIILVNTGSSGGLHH1906 pKa = 4.62VTGDD1910 pKa = 3.56GSTAASGGTIANKK1923 pKa = 9.4TGADD1927 pKa = 3.6GSSTQGTGVFLNNTHH1942 pKa = 6.63DD1943 pKa = 3.7VEE1945 pKa = 4.24LAYY1948 pKa = 9.74MQLNDD1953 pKa = 4.08FSNFAIRR1960 pKa = 11.84GNNVTGFTLDD1970 pKa = 3.28HH1971 pKa = 6.42SVINGDD1977 pKa = 3.4NGTSNSLSDD1986 pKa = 4.16PLASFANVGEE1996 pKa = 4.08DD1997 pKa = 4.02AIRR2000 pKa = 11.84FTNLLGSGSITNSTISGGYY2019 pKa = 7.68TNTIMVEE2026 pKa = 4.08NNTGILNRR2034 pKa = 11.84LTIDD2038 pKa = 2.88HH2039 pKa = 6.08STIGGRR2045 pKa = 11.84TDD2047 pKa = 3.3NGTGMNDD2054 pKa = 4.35GINFQADD2061 pKa = 3.45TGSTAMNLTVTNSALTTARR2080 pKa = 11.84ANLIAVTNQTGTQTDD2095 pKa = 3.56VKK2097 pKa = 10.59FDD2099 pKa = 4.04NNTLTNNHH2107 pKa = 7.2PLTVTGGTAGDD2118 pKa = 3.79FAGLGGMTFDD2128 pKa = 4.5ISNNTFDD2135 pKa = 3.37MHH2137 pKa = 6.8AANSGTGIKK2146 pKa = 10.67SNVIEE2151 pKa = 4.22VNKK2154 pKa = 10.48GSGSGSSTFDD2164 pKa = 3.06GTISGNTIGVAATAHH2179 pKa = 5.95SGSGIGANDD2188 pKa = 3.33IDD2190 pKa = 5.81INSQDD2195 pKa = 3.26NGIFNILVQNNILTHH2210 pKa = 6.51YY2211 pKa = 7.77DD2212 pKa = 3.03TAGIQISQISGSSTVNATVIGNTTSNGDD2240 pKa = 3.54SNAFAGLYY2248 pKa = 9.28VVAGSGLATDD2258 pKa = 3.86NGVVNLKK2265 pKa = 10.65VGGTGTEE2272 pKa = 3.9RR2273 pKa = 11.84NDD2275 pKa = 3.9FSNGDD2280 pKa = 3.82PANGSDD2286 pKa = 4.17VFLQQSSPSSTSVFNLTGSPATNNATLAAITASIQANNLNPGTTTVGTSGTINLVSTTPTLPLLAAPGGVQASSPTTGEE2365 pKa = 3.59MHH2367 pKa = 6.66LTQSEE2372 pKa = 4.33LDD2374 pKa = 3.9SVVAAAIAQWAAAGASASQLAALHH2398 pKa = 6.26AATFSVADD2406 pKa = 4.11LSGSIIGDD2414 pKa = 3.49EE2415 pKa = 4.3SSPAHH2420 pKa = 5.69ITIDD2424 pKa = 3.17TDD2426 pKa = 3.62AAGHH2430 pKa = 6.07GWFIDD2435 pKa = 3.63PTPSDD2440 pKa = 3.4NSEE2443 pKa = 3.77FTHH2446 pKa = 6.13AQNAAGTDD2454 pKa = 3.83LLTDD2458 pKa = 3.89PTNAAAGHH2466 pKa = 7.0LDD2468 pKa = 3.44LLTTVSHH2475 pKa = 6.2EE2476 pKa = 4.16MGHH2479 pKa = 5.95VLGLPDD2485 pKa = 3.85STSPSAVNDD2494 pKa = 3.38LMYY2497 pKa = 11.04VSLVDD2502 pKa = 3.55GEE2504 pKa = 4.58RR2505 pKa = 11.84RR2506 pKa = 11.84LPDD2509 pKa = 3.77AADD2512 pKa = 3.33VAQANGSTFSFQAVGTTAPVVTAAPVLDD2540 pKa = 4.74AGHH2543 pKa = 6.87GGGTLVGTAGADD2555 pKa = 3.02NFVFANVDD2563 pKa = 3.35VHH2565 pKa = 6.86GSAPPPITHH2574 pKa = 6.18VANYY2578 pKa = 10.42SFAQGDD2584 pKa = 3.8TFDD2587 pKa = 4.95FSVLTSQLHH2596 pKa = 5.53GSGVSDD2602 pKa = 3.65ALVVRR2607 pKa = 11.84AIEE2610 pKa = 4.17GAGGSFATLQVNTADD2625 pKa = 3.43SSMGSKK2631 pKa = 10.3AGSNWVSVAQIDD2643 pKa = 4.14GAHH2646 pKa = 6.76AGDD2649 pKa = 4.15ALNVLVDD2656 pKa = 3.27THH2658 pKa = 6.34SAVHH2662 pKa = 6.68ASQIHH2667 pKa = 5.74VDD2669 pKa = 3.86LLVV2672 pKa = 3.37
MM1 pKa = 6.89STIPAFTFGGITRR14 pKa = 11.84PALILDD20 pKa = 3.7PSGNIILDD28 pKa = 3.51PAAQAFLDD36 pKa = 4.13TYY38 pKa = 8.75GTKK41 pKa = 10.45ALYY44 pKa = 10.61AGVPPGTHH52 pKa = 6.69FPPIPDD58 pKa = 4.32LLTTPVDD65 pKa = 3.88TNGTANSVAEE75 pKa = 4.3GAAVNTLVGITASSHH90 pKa = 5.49SLLPFSITYY99 pKa = 10.27SLSADD104 pKa = 3.43SSSGGFKK111 pKa = 9.66IDD113 pKa = 3.28PTTGVVSVANSAKK126 pKa = 9.68IDD128 pKa = 3.82FEE130 pKa = 4.5SSPGHH135 pKa = 6.85AYY137 pKa = 8.76TVTVQATDD145 pKa = 4.6GIFTTSQNFTIGVSDD160 pKa = 3.95VAPSTPVDD168 pKa = 3.49GNAATNTIAEE178 pKa = 4.44GAAVGTAVGVTAHH191 pKa = 6.0STDD194 pKa = 3.34INGGTVTYY202 pKa = 10.6SLTGDD207 pKa = 3.4TSGGGFTVNAATGVVTVADD226 pKa = 3.81STKK229 pKa = 9.92IDD231 pKa = 3.79YY232 pKa = 7.64EE233 pKa = 4.35TAPGHH238 pKa = 6.26AYY240 pKa = 10.39SVTVQASDD248 pKa = 3.46GTLTSATQTFTIAVSDD264 pKa = 4.15VPLPTPADD272 pKa = 3.69TNPAANQVAEE282 pKa = 4.45GAAAGTLVGLTATANDD298 pKa = 4.21PNGPTTTYY306 pKa = 11.34SLIGDD311 pKa = 4.04TSGGGFTIDD320 pKa = 3.51PATGVVTVADD330 pKa = 3.69PAKK333 pKa = 10.28IDD335 pKa = 3.59FEE337 pKa = 4.64SAAGHH342 pKa = 6.6AYY344 pKa = 10.33SVTVKK349 pKa = 10.79ADD351 pKa = 3.7DD352 pKa = 4.45GLASTTQTFSIAVTDD367 pKa = 3.99VAPSAPTDD375 pKa = 3.45SDD377 pKa = 3.62ASANTIAEE385 pKa = 4.28GAANGSTVGITAHH398 pKa = 5.68STDD401 pKa = 3.41VNGPAVTYY409 pKa = 10.67SLVGDD414 pKa = 3.99TSGGGFTINAATGVITVADD433 pKa = 3.82STKK436 pKa = 9.89IDD438 pKa = 3.83YY439 pKa = 7.68EE440 pKa = 4.29TSPGHH445 pKa = 6.9AYY447 pKa = 8.8TVTAQASDD455 pKa = 3.54GTLASSQTFTIGVTDD470 pKa = 4.06VPPSNPVDD478 pKa = 3.84SNAAANAVAEE488 pKa = 4.54GAAAGATVGITASSTDD504 pKa = 3.35VNGPAVTYY512 pKa = 10.68SLTGDD517 pKa = 3.4TSGGGFTINSATGVVTVADD536 pKa = 3.88PTKK539 pKa = 10.0IDD541 pKa = 3.74YY542 pKa = 9.46EE543 pKa = 4.44SSPGHH548 pKa = 6.67AYY550 pKa = 10.64SITVQSSDD558 pKa = 3.08GTLTSSQVFTIGVNDD573 pKa = 3.8VAPSTPVDD581 pKa = 3.52SDD583 pKa = 3.19ATANAVAEE591 pKa = 4.49GAAAGTNVGITAHH604 pKa = 6.25SVDD607 pKa = 3.78VNGPAVTYY615 pKa = 10.67SLVGDD620 pKa = 3.99TSGGGFTINSATGVVTVADD639 pKa = 3.56PTKK642 pKa = 10.37IDD644 pKa = 3.84FEE646 pKa = 4.89SSGGSYY652 pKa = 9.14TVVAQSTDD660 pKa = 3.12GTLTNSQAFVIAVTDD675 pKa = 3.77VAPSTPVDD683 pKa = 3.74SNAAANTIAEE693 pKa = 4.36GAAAGSTVGITASSTDD709 pKa = 3.35VNGPAVTYY717 pKa = 10.67SLVGDD722 pKa = 3.99TSGGGFTINATTGVVTVTDD741 pKa = 3.77PTKK744 pKa = 10.13IDD746 pKa = 3.54YY747 pKa = 10.42EE748 pKa = 4.44SAPGHH753 pKa = 6.79AYY755 pKa = 8.84TVTSQASDD763 pKa = 3.4GTLTSSQAFTIAVTDD778 pKa = 3.6VAPSVPVDD786 pKa = 3.31SDD788 pKa = 3.42AAANAVTEE796 pKa = 4.16GAPAGSTVGVTASSTDD812 pKa = 3.38VNGPPVTYY820 pKa = 10.49SLTGDD825 pKa = 3.4TSGGGFTINATTGVVTVADD844 pKa = 4.16PSKK847 pKa = 10.46IDD849 pKa = 3.47YY850 pKa = 10.22EE851 pKa = 4.48SAPGHH856 pKa = 6.4AYY858 pKa = 10.34SVTVTSSDD866 pKa = 2.82GTMANSQVFTIAVNDD881 pKa = 4.12AAPSTPVDD889 pKa = 3.27SDD891 pKa = 3.44AAANTVVEE899 pKa = 4.72GAAAGTTVGVTASSTDD915 pKa = 3.33VNGPAVTYY923 pKa = 10.68SLTGDD928 pKa = 3.4TSGGGFTINAATGVVTVADD947 pKa = 3.56PTKK950 pKa = 10.4IDD952 pKa = 4.04FEE954 pKa = 4.72STAPGHH960 pKa = 6.27TYY962 pKa = 9.93TVTATASDD970 pKa = 3.79GTLASSQIFTIAVTDD985 pKa = 4.47APPSTPVDD993 pKa = 3.16SDD995 pKa = 3.69GAINQVSQNAPVGTYY1010 pKa = 9.26TGLTASSTDD1019 pKa = 3.33VNGPAVTYY1027 pKa = 10.67SLVGDD1032 pKa = 3.97TSGGGFAIDD1041 pKa = 3.44ASTGKK1046 pKa = 8.5VTVADD1051 pKa = 3.61STKK1054 pKa = 9.96IPFNAGSPSFDD1065 pKa = 3.15VTVQSSDD1072 pKa = 3.39GTDD1075 pKa = 2.98TSQQTFTINVIQNTPPVANDD1095 pKa = 3.78DD1096 pKa = 4.05SLSATEE1102 pKa = 5.61AGGLNNSIPGSNPSGNVILGTGSAGAVQDD1131 pKa = 4.3TDD1133 pKa = 3.73AQDD1136 pKa = 3.5PSTALTVVAVHH1147 pKa = 6.6TGPEE1151 pKa = 4.29GGSTGTGTVGSPIVGLHH1168 pKa = 5.31GTLTLDD1174 pKa = 3.06SHH1176 pKa = 6.64GAYY1179 pKa = 9.26TYY1181 pKa = 9.67TVNQTDD1187 pKa = 3.69PTVQALHH1194 pKa = 6.12TAAEE1198 pKa = 4.5TITDD1202 pKa = 3.56QFNYY1206 pKa = 9.51TIQDD1210 pKa = 3.32TGGLQDD1216 pKa = 3.34TATITFTIHH1225 pKa = 6.7GADD1228 pKa = 5.0DD1229 pKa = 4.41LPQAIADD1236 pKa = 3.96TGTMTEE1242 pKa = 4.78DD1243 pKa = 4.38DD1244 pKa = 4.91APTLFNVLANDD1255 pKa = 4.09TLDD1258 pKa = 4.45PDD1260 pKa = 3.68HH1261 pKa = 6.8TALNTIAVGPGSITVTGPAGEE1282 pKa = 4.5TFANTDD1288 pKa = 2.74ATATVVSNEE1297 pKa = 4.09VQVSLNNAHH1306 pKa = 6.32FQQLAVGEE1314 pKa = 4.55HH1315 pKa = 5.31ATITVPYY1322 pKa = 9.14TLTGDD1327 pKa = 3.43TGEE1330 pKa = 4.22TSTANLVVTVNGVNDD1345 pKa = 3.47VPVAVDD1351 pKa = 3.86DD1352 pKa = 4.35SGSMTEE1358 pKa = 4.51DD1359 pKa = 3.27QSGHH1363 pKa = 6.24AFTVLSNDD1371 pKa = 3.74TLDD1374 pKa = 4.77PDD1376 pKa = 3.82HH1377 pKa = 6.9GAPNNVTAGAVTNLSAPAGEE1397 pKa = 5.37GITTADD1403 pKa = 2.8IGVSVNASNQVVVNLGANFQHH1424 pKa = 6.09MQDD1427 pKa = 3.67GQTATFDD1434 pKa = 3.48VAYY1437 pKa = 8.76TLHH1440 pKa = 7.18GDD1442 pKa = 3.7QPGDD1446 pKa = 3.12TSTAEE1451 pKa = 3.77LHH1453 pKa = 5.15VTVTGVNDD1461 pKa = 3.53APVAGNFTFNGVNSATGNTDD1481 pKa = 5.28LIVNDD1486 pKa = 4.46PATTAITEE1494 pKa = 4.24TTPNKK1499 pKa = 9.61TISASLLSGASDD1511 pKa = 3.43VDD1513 pKa = 3.92GPGPLTVVAGTIATAHH1529 pKa = 6.31GGSVTMQSDD1538 pKa = 3.47GDD1540 pKa = 4.34FIYY1543 pKa = 10.36KK1544 pKa = 10.23PPVGYY1549 pKa = 8.84TGSDD1553 pKa = 3.13SFNYY1557 pKa = 9.32QVSDD1561 pKa = 3.6QNTGASGVGLGTGTVTINVAAPKK1584 pKa = 9.48VWYY1587 pKa = 10.41VNADD1591 pKa = 3.18AATDD1595 pKa = 3.83GDD1597 pKa = 4.39GSSEE1601 pKa = 4.09HH1602 pKa = 6.91PFNNLTHH1609 pKa = 6.67FNGAGGVDD1617 pKa = 3.68GAGDD1621 pKa = 4.07TIVLEE1626 pKa = 4.48TATAHH1631 pKa = 4.53YY1632 pKa = 9.47TGGLTLEE1639 pKa = 4.48NNEE1642 pKa = 3.97QLISQSAGVTINGTTLFAASGANAVVDD1669 pKa = 3.87GGVVLGSGNTIEE1681 pKa = 5.24GVDD1684 pKa = 4.19FGTTSGFAVSGSSVGTLHH1702 pKa = 7.37LNDD1705 pKa = 3.32THH1707 pKa = 6.65TGVINNSSGGGISIGGASNVLNVDD1731 pKa = 4.61FNSITTGGGTNGIALTNSSGTFHH1754 pKa = 7.02AHH1756 pKa = 6.79GGTISNASGADD1767 pKa = 3.29VALNGGTVTFTDD1779 pKa = 4.46DD1780 pKa = 3.69GAISDD1785 pKa = 3.86STGTVVSVANMTGNTQAFTGAIGTTGSTDD1814 pKa = 3.1GAIALSSNTGATINFSGGMALSTGASDD1841 pKa = 4.31AFSATGGGTVNVTGTNHH1858 pKa = 6.39LTTTTGTALNISNTTIGSSNVTFHH1882 pKa = 7.67DD1883 pKa = 3.37ISSNGGSSNGIILVNTGSSGGLHH1906 pKa = 4.62VTGDD1910 pKa = 3.56GSTAASGGTIANKK1923 pKa = 9.4TGADD1927 pKa = 3.6GSSTQGTGVFLNNTHH1942 pKa = 6.63DD1943 pKa = 3.7VEE1945 pKa = 4.24LAYY1948 pKa = 9.74MQLNDD1953 pKa = 4.08FSNFAIRR1960 pKa = 11.84GNNVTGFTLDD1970 pKa = 3.28HH1971 pKa = 6.42SVINGDD1977 pKa = 3.4NGTSNSLSDD1986 pKa = 4.16PLASFANVGEE1996 pKa = 4.08DD1997 pKa = 4.02AIRR2000 pKa = 11.84FTNLLGSGSITNSTISGGYY2019 pKa = 7.68TNTIMVEE2026 pKa = 4.08NNTGILNRR2034 pKa = 11.84LTIDD2038 pKa = 2.88HH2039 pKa = 6.08STIGGRR2045 pKa = 11.84TDD2047 pKa = 3.3NGTGMNDD2054 pKa = 4.35GINFQADD2061 pKa = 3.45TGSTAMNLTVTNSALTTARR2080 pKa = 11.84ANLIAVTNQTGTQTDD2095 pKa = 3.56VKK2097 pKa = 10.59FDD2099 pKa = 4.04NNTLTNNHH2107 pKa = 7.2PLTVTGGTAGDD2118 pKa = 3.79FAGLGGMTFDD2128 pKa = 4.5ISNNTFDD2135 pKa = 3.37MHH2137 pKa = 6.8AANSGTGIKK2146 pKa = 10.67SNVIEE2151 pKa = 4.22VNKK2154 pKa = 10.48GSGSGSSTFDD2164 pKa = 3.06GTISGNTIGVAATAHH2179 pKa = 5.95SGSGIGANDD2188 pKa = 3.33IDD2190 pKa = 5.81INSQDD2195 pKa = 3.26NGIFNILVQNNILTHH2210 pKa = 6.51YY2211 pKa = 7.77DD2212 pKa = 3.03TAGIQISQISGSSTVNATVIGNTTSNGDD2240 pKa = 3.54SNAFAGLYY2248 pKa = 9.28VVAGSGLATDD2258 pKa = 3.86NGVVNLKK2265 pKa = 10.65VGGTGTEE2272 pKa = 3.9RR2273 pKa = 11.84NDD2275 pKa = 3.9FSNGDD2280 pKa = 3.82PANGSDD2286 pKa = 4.17VFLQQSSPSSTSVFNLTGSPATNNATLAAITASIQANNLNPGTTTVGTSGTINLVSTTPTLPLLAAPGGVQASSPTTGEE2365 pKa = 3.59MHH2367 pKa = 6.66LTQSEE2372 pKa = 4.33LDD2374 pKa = 3.9SVVAAAIAQWAAAGASASQLAALHH2398 pKa = 6.26AATFSVADD2406 pKa = 4.11LSGSIIGDD2414 pKa = 3.49EE2415 pKa = 4.3SSPAHH2420 pKa = 5.69ITIDD2424 pKa = 3.17TDD2426 pKa = 3.62AAGHH2430 pKa = 6.07GWFIDD2435 pKa = 3.63PTPSDD2440 pKa = 3.4NSEE2443 pKa = 3.77FTHH2446 pKa = 6.13AQNAAGTDD2454 pKa = 3.83LLTDD2458 pKa = 3.89PTNAAAGHH2466 pKa = 7.0LDD2468 pKa = 3.44LLTTVSHH2475 pKa = 6.2EE2476 pKa = 4.16MGHH2479 pKa = 5.95VLGLPDD2485 pKa = 3.85STSPSAVNDD2494 pKa = 3.38LMYY2497 pKa = 11.04VSLVDD2502 pKa = 3.55GEE2504 pKa = 4.58RR2505 pKa = 11.84RR2506 pKa = 11.84LPDD2509 pKa = 3.77AADD2512 pKa = 3.33VAQANGSTFSFQAVGTTAPVVTAAPVLDD2540 pKa = 4.74AGHH2543 pKa = 6.87GGGTLVGTAGADD2555 pKa = 3.02NFVFANVDD2563 pKa = 3.35VHH2565 pKa = 6.86GSAPPPITHH2574 pKa = 6.18VANYY2578 pKa = 10.42SFAQGDD2584 pKa = 3.8TFDD2587 pKa = 4.95FSVLTSQLHH2596 pKa = 5.53GSGVSDD2602 pKa = 3.65ALVVRR2607 pKa = 11.84AIEE2610 pKa = 4.17GAGGSFATLQVNTADD2625 pKa = 3.43SSMGSKK2631 pKa = 10.3AGSNWVSVAQIDD2643 pKa = 4.14GAHH2646 pKa = 6.76AGDD2649 pKa = 4.15ALNVLVDD2656 pKa = 3.27THH2658 pKa = 6.34SAVHH2662 pKa = 6.68ASQIHH2667 pKa = 5.74VDD2669 pKa = 3.86LLVV2672 pKa = 3.37
Molecular weight: 263.28 kDa
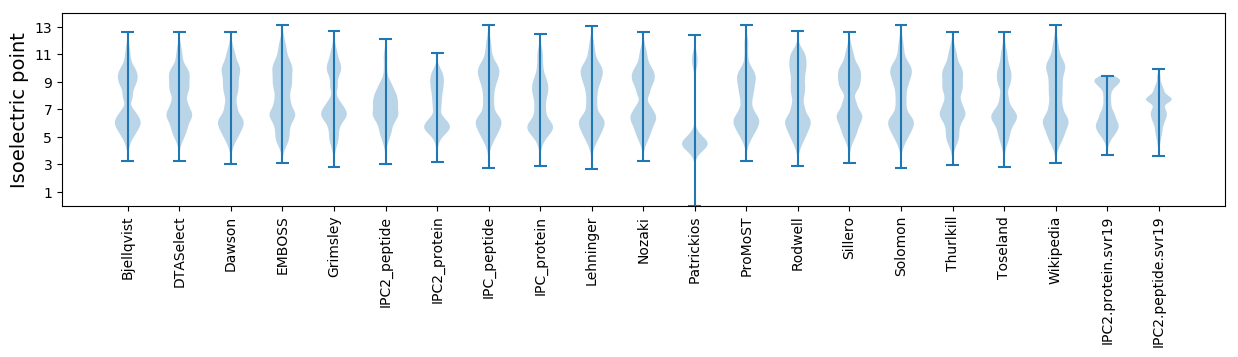
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N6ENP2|A0A1N6ENP2_9BRAD Uncharacterized conserved protein contains HEPN domain OS=Bradyrhizobium erythrophlei OX=1437360 GN=SAMN05443247_00159 PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 10.25VRR4 pKa = 11.84NSLKK8 pKa = 10.15SLRR11 pKa = 11.84GRR13 pKa = 11.84HH14 pKa = 5.06RR15 pKa = 11.84NNRR18 pKa = 11.84LVRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 10.2GRR25 pKa = 11.84VYY27 pKa = 10.74VINKK31 pKa = 4.67VQRR34 pKa = 11.84RR35 pKa = 11.84FKK37 pKa = 10.83ARR39 pKa = 11.84QGG41 pKa = 3.34
MM1 pKa = 7.65KK2 pKa = 10.25VRR4 pKa = 11.84NSLKK8 pKa = 10.15SLRR11 pKa = 11.84GRR13 pKa = 11.84HH14 pKa = 5.06RR15 pKa = 11.84NNRR18 pKa = 11.84LVRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 10.2GRR25 pKa = 11.84VYY27 pKa = 10.74VINKK31 pKa = 4.67VQRR34 pKa = 11.84RR35 pKa = 11.84FKK37 pKa = 10.83ARR39 pKa = 11.84QGG41 pKa = 3.34
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2999090 |
26 |
3139 |
282.8 |
30.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.093 ± 0.032 | 0.945 ± 0.008 |
5.415 ± 0.019 | 5.273 ± 0.022 |
3.826 ± 0.015 | 8.155 ± 0.026 |
2.113 ± 0.012 | 5.425 ± 0.017 |
3.751 ± 0.023 | 9.814 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.43 ± 0.012 | 2.854 ± 0.018 |
5.32 ± 0.019 | 3.232 ± 0.015 |
7.144 ± 0.033 | 5.773 ± 0.02 |
5.378 ± 0.025 | 7.344 ± 0.02 |
1.403 ± 0.01 | 2.31 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
