
Gordonia phage Wizard
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Wizardvirus; Gordonia virus Wizard
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
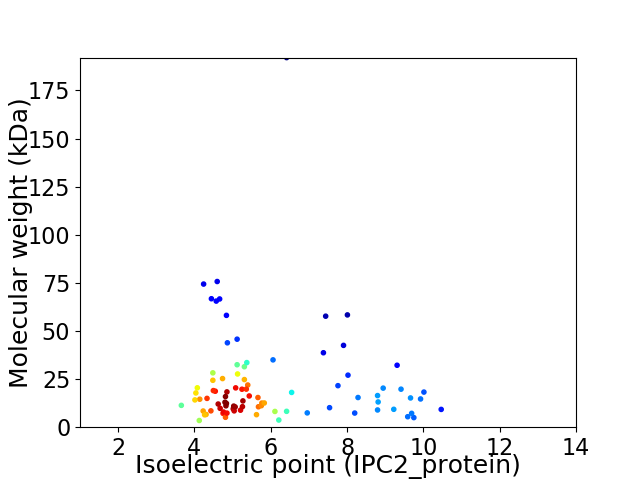
Virtual 2D-PAGE plot for 89 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
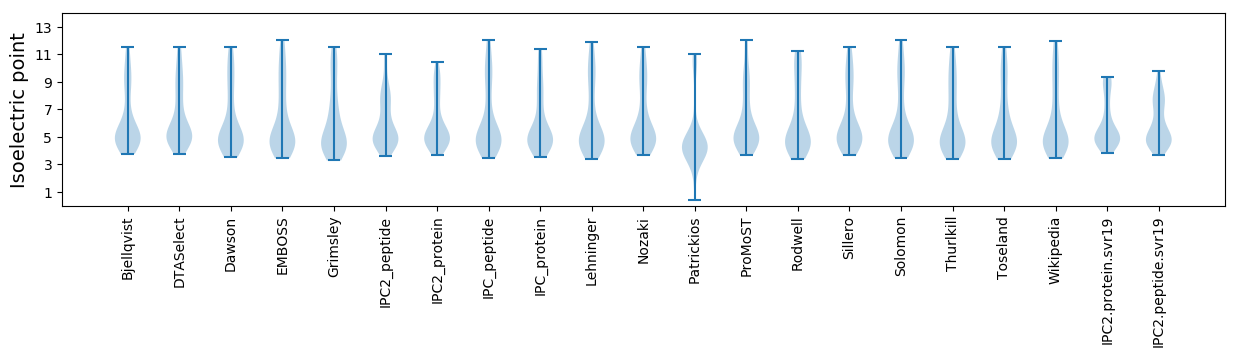
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160DCM5|A0A160DCM5_9CAUD Uncharacterized protein OS=Gordonia phage Wizard OX=1838083 GN=74 PE=4 SV=1
MM1 pKa = 7.26IRR3 pKa = 11.84KK4 pKa = 9.67LSTALAALAILAAPVAVAMANPTPAPAQVPAPCVIQLGNGGTAPCPPPIVSTDD57 pKa = 3.04GSSIGGGANTDD68 pKa = 3.75PVNMGDD74 pKa = 4.63LPRR77 pKa = 11.84TGPDD81 pKa = 3.25YY82 pKa = 11.07SYY84 pKa = 10.78EE85 pKa = 4.21APTWTPDD92 pKa = 3.15EE93 pKa = 4.69EE94 pKa = 4.7EE95 pKa = 5.07PDD97 pKa = 3.7TEE99 pKa = 4.46EE100 pKa = 3.96PTEE103 pKa = 4.21TEE105 pKa = 4.06EE106 pKa = 4.15PAPEE110 pKa = 4.82AEE112 pKa = 4.19
MM1 pKa = 7.26IRR3 pKa = 11.84KK4 pKa = 9.67LSTALAALAILAAPVAVAMANPTPAPAQVPAPCVIQLGNGGTAPCPPPIVSTDD57 pKa = 3.04GSSIGGGANTDD68 pKa = 3.75PVNMGDD74 pKa = 4.63LPRR77 pKa = 11.84TGPDD81 pKa = 3.25YY82 pKa = 11.07SYY84 pKa = 10.78EE85 pKa = 4.21APTWTPDD92 pKa = 3.15EE93 pKa = 4.69EE94 pKa = 4.7EE95 pKa = 5.07PDD97 pKa = 3.7TEE99 pKa = 4.46EE100 pKa = 3.96PTEE103 pKa = 4.21TEE105 pKa = 4.06EE106 pKa = 4.15PAPEE110 pKa = 4.82AEE112 pKa = 4.19
Molecular weight: 11.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166Y2H9|A0A166Y2H9_9CAUD HTH DNA binding domain protein OS=Gordonia phage Wizard OX=1838083 GN=48 PE=4 SV=1
MM1 pKa = 7.19VNNIVHH7 pKa = 5.34MTNMRR12 pKa = 11.84NTGKK16 pKa = 10.64AKK18 pKa = 10.09FVPASWQPLIAGFLRR33 pKa = 11.84HH34 pKa = 5.95LRR36 pKa = 11.84AAGQPDD42 pKa = 3.68TTINTRR48 pKa = 11.84RR49 pKa = 11.84QQIEE53 pKa = 3.81RR54 pKa = 11.84MARR57 pKa = 11.84ALEE60 pKa = 4.22GEE62 pKa = 4.21PAEE65 pKa = 4.35VTRR68 pKa = 11.84DD69 pKa = 3.58AITDD73 pKa = 3.16YY74 pKa = 11.03HH75 pKa = 7.2AAQTWALEE83 pKa = 3.96TRR85 pKa = 11.84RR86 pKa = 11.84GHH88 pKa = 6.45RR89 pKa = 11.84NAAVSFFAWAHH100 pKa = 5.07SCGHH104 pKa = 6.14IPEE107 pKa = 5.11NPAQHH112 pKa = 6.34LPIVKK117 pKa = 10.05SSVPAPRR124 pKa = 11.84PAPDD128 pKa = 2.72RR129 pKa = 11.84VVNVALKK136 pKa = 9.37TPEE139 pKa = 3.91KK140 pKa = 10.24RR141 pKa = 11.84IQLMLRR147 pKa = 11.84LSCEE151 pKa = 3.66LGLRR155 pKa = 11.84RR156 pKa = 11.84AEE158 pKa = 4.07VAVVSTTDD166 pKa = 3.16VEE168 pKa = 4.77DD169 pKa = 4.3GVGGAMLRR177 pKa = 11.84VHH179 pKa = 6.86GKK181 pKa = 8.84GGKK184 pKa = 9.04IRR186 pKa = 11.84VIPISDD192 pKa = 3.53EE193 pKa = 3.66LALRR197 pKa = 11.84IEE199 pKa = 5.04DD200 pKa = 3.63GAAGHH205 pKa = 6.29TPGAPRR211 pKa = 11.84TGYY214 pKa = 10.61LFPGNDD220 pKa = 3.86GGHH223 pKa = 7.34LSPRR227 pKa = 11.84WVGTLCAEE235 pKa = 4.6ALPGGWTMHH244 pKa = 5.65TLRR247 pKa = 11.84HH248 pKa = 5.69RR249 pKa = 11.84FATRR253 pKa = 11.84AYY255 pKa = 9.41RR256 pKa = 11.84GSRR259 pKa = 11.84NLRR262 pKa = 11.84AVQVLLGHH270 pKa = 7.11ASLATTQRR278 pKa = 11.84YY279 pKa = 5.49TAVDD283 pKa = 3.57EE284 pKa = 4.78DD285 pKa = 4.18EE286 pKa = 4.97VRR288 pKa = 11.84AAMNSAAA295 pKa = 4.64
MM1 pKa = 7.19VNNIVHH7 pKa = 5.34MTNMRR12 pKa = 11.84NTGKK16 pKa = 10.64AKK18 pKa = 10.09FVPASWQPLIAGFLRR33 pKa = 11.84HH34 pKa = 5.95LRR36 pKa = 11.84AAGQPDD42 pKa = 3.68TTINTRR48 pKa = 11.84RR49 pKa = 11.84QQIEE53 pKa = 3.81RR54 pKa = 11.84MARR57 pKa = 11.84ALEE60 pKa = 4.22GEE62 pKa = 4.21PAEE65 pKa = 4.35VTRR68 pKa = 11.84DD69 pKa = 3.58AITDD73 pKa = 3.16YY74 pKa = 11.03HH75 pKa = 7.2AAQTWALEE83 pKa = 3.96TRR85 pKa = 11.84RR86 pKa = 11.84GHH88 pKa = 6.45RR89 pKa = 11.84NAAVSFFAWAHH100 pKa = 5.07SCGHH104 pKa = 6.14IPEE107 pKa = 5.11NPAQHH112 pKa = 6.34LPIVKK117 pKa = 10.05SSVPAPRR124 pKa = 11.84PAPDD128 pKa = 2.72RR129 pKa = 11.84VVNVALKK136 pKa = 9.37TPEE139 pKa = 3.91KK140 pKa = 10.24RR141 pKa = 11.84IQLMLRR147 pKa = 11.84LSCEE151 pKa = 3.66LGLRR155 pKa = 11.84RR156 pKa = 11.84AEE158 pKa = 4.07VAVVSTTDD166 pKa = 3.16VEE168 pKa = 4.77DD169 pKa = 4.3GVGGAMLRR177 pKa = 11.84VHH179 pKa = 6.86GKK181 pKa = 8.84GGKK184 pKa = 9.04IRR186 pKa = 11.84VIPISDD192 pKa = 3.53EE193 pKa = 3.66LALRR197 pKa = 11.84IEE199 pKa = 5.04DD200 pKa = 3.63GAAGHH205 pKa = 6.29TPGAPRR211 pKa = 11.84TGYY214 pKa = 10.61LFPGNDD220 pKa = 3.86GGHH223 pKa = 7.34LSPRR227 pKa = 11.84WVGTLCAEE235 pKa = 4.6ALPGGWTMHH244 pKa = 5.65TLRR247 pKa = 11.84HH248 pKa = 5.69RR249 pKa = 11.84FATRR253 pKa = 11.84AYY255 pKa = 9.41RR256 pKa = 11.84GSRR259 pKa = 11.84NLRR262 pKa = 11.84AVQVLLGHH270 pKa = 7.11ASLATTQRR278 pKa = 11.84YY279 pKa = 5.49TAVDD283 pKa = 3.57EE284 pKa = 4.78DD285 pKa = 4.18EE286 pKa = 4.97VRR288 pKa = 11.84AAMNSAAA295 pKa = 4.64
Molecular weight: 32.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
18513 |
29 |
1843 |
208.0 |
22.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.37 ± 0.528 | 0.832 ± 0.134 |
7.163 ± 0.325 | 5.634 ± 0.343 |
2.485 ± 0.173 | 8.643 ± 0.484 |
2.15 ± 0.207 | 4.202 ± 0.203 |
2.825 ± 0.25 | 7.832 ± 0.251 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.069 ± 0.129 | 2.793 ± 0.174 |
6.293 ± 0.34 | 3.354 ± 0.16 |
7.298 ± 0.415 | 5.153 ± 0.227 |
7.119 ± 0.335 | 7.562 ± 0.212 |
2.209 ± 0.183 | 2.015 ± 0.152 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
