
Desulfotignum phosphitoxidans DSM 13687
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfotignum; Desulfotignum phosphitoxidans
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
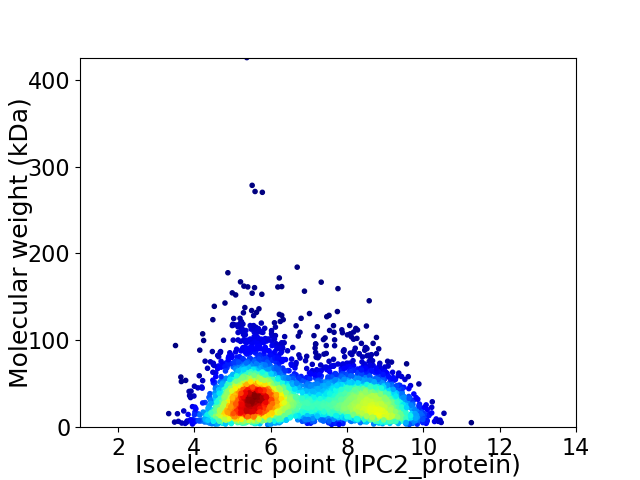
Virtual 2D-PAGE plot for 4515 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S0G4F0|S0G4F0_9DELT Excisionase DNA binding family protein OS=Desulfotignum phosphitoxidans DSM 13687 OX=1286635 GN=Dpo_5c01100 PE=4 SV=1
MM1 pKa = 7.9PLPGFDD7 pKa = 3.2KK8 pKa = 10.75AYY10 pKa = 10.42YY11 pKa = 9.87LQAKK15 pKa = 7.96LTALQTTLPEE25 pKa = 4.27WVGKK29 pKa = 6.57TTTDD33 pKa = 4.76LEE35 pKa = 4.53TLLTNQNFTAEE46 pKa = 4.08SHH48 pKa = 5.37YY49 pKa = 10.92QLYY52 pKa = 10.55GYY54 pKa = 9.69QEE56 pKa = 4.2GLAPNAFFNADD67 pKa = 3.53EE68 pKa = 4.18YY69 pKa = 9.87TQAKK73 pKa = 8.41AVSMFNAGGYY83 pKa = 7.52STISDD88 pKa = 2.99ARR90 pKa = 11.84AAFVAAWTGDD100 pKa = 3.54VYY102 pKa = 11.72NHH104 pKa = 5.88YY105 pKa = 10.04LQYY108 pKa = 11.17GSQEE112 pKa = 4.38GINPSNAFDD121 pKa = 3.53EE122 pKa = 4.48SSYY125 pKa = 10.69YY126 pKa = 10.57ASKK129 pKa = 10.77LADD132 pKa = 3.66LQANADD138 pKa = 4.05TSAEE142 pKa = 3.99WATKK146 pKa = 10.28SVAEE150 pKa = 4.46LKK152 pKa = 10.99AFFNAQGFSALHH164 pKa = 6.47HH165 pKa = 5.94YY166 pKa = 9.72NVYY169 pKa = 10.4GAGEE173 pKa = 5.04GIPVTEE179 pKa = 4.39VPVSEE184 pKa = 4.45AVNSEE189 pKa = 3.57TDD191 pKa = 3.7EE192 pKa = 4.42IDD194 pKa = 3.4EE195 pKa = 4.32TDD197 pKa = 3.52EE198 pKa = 4.04TDD200 pKa = 3.39EE201 pKa = 4.84TGGTPAKK208 pKa = 10.47GMNFQLTTNDD218 pKa = 3.46DD219 pKa = 3.98SFTGGAANDD228 pKa = 4.12HH229 pKa = 6.87FYY231 pKa = 11.36GIYY234 pKa = 10.69DD235 pKa = 3.69GVTFLAQNTFNSGDD249 pKa = 3.62ILDD252 pKa = 4.59GAGGTDD258 pKa = 3.28TLHH261 pKa = 6.66INHH264 pKa = 6.42FEE266 pKa = 4.11DD267 pKa = 4.34VAIDD271 pKa = 4.32LPDD274 pKa = 3.89TLWTGISGIEE284 pKa = 4.21NIVMSTTGSGAHH296 pKa = 7.16DD297 pKa = 5.35IITGTNFNTAFASGGVDD314 pKa = 3.15LWTATTGAGAITIDD328 pKa = 3.48MASFTGTAKK337 pKa = 9.13VTADD341 pKa = 3.34SAAGAIVLSGVKK353 pKa = 10.31FSQIDD358 pKa = 3.56VTTTGSGAQTITSTGASDD376 pKa = 4.08VIVNATSSEE385 pKa = 4.1GATSITTGAGDD396 pKa = 3.69DD397 pKa = 4.38TIALHH402 pKa = 5.87ATSADD407 pKa = 3.28GLNTITAGAGADD419 pKa = 4.03TINLYY424 pKa = 11.39ANFASIDD431 pKa = 3.92KK432 pKa = 10.29IVQADD437 pKa = 4.1GDD439 pKa = 4.64SMASTANTITSNIATGDD456 pKa = 3.65TITFGNGLDD465 pKa = 3.13IVRR468 pKa = 11.84NFGVNDD474 pKa = 4.05NIDD477 pKa = 3.61VGTGGAAVSGLTMDD491 pKa = 4.39KK492 pKa = 11.14ADD494 pKa = 4.09FAATTTIFLSGTYY507 pKa = 10.43AEE509 pKa = 4.61DD510 pKa = 3.66TFTIAADD517 pKa = 3.66GGGTDD522 pKa = 3.99TLLLDD527 pKa = 3.81TTAVDD532 pKa = 3.87DD533 pKa = 4.27TDD535 pKa = 3.85IATADD540 pKa = 3.24TWILLSGVTSDD551 pKa = 4.2ALSAGSFVV559 pKa = 3.47
MM1 pKa = 7.9PLPGFDD7 pKa = 3.2KK8 pKa = 10.75AYY10 pKa = 10.42YY11 pKa = 9.87LQAKK15 pKa = 7.96LTALQTTLPEE25 pKa = 4.27WVGKK29 pKa = 6.57TTTDD33 pKa = 4.76LEE35 pKa = 4.53TLLTNQNFTAEE46 pKa = 4.08SHH48 pKa = 5.37YY49 pKa = 10.92QLYY52 pKa = 10.55GYY54 pKa = 9.69QEE56 pKa = 4.2GLAPNAFFNADD67 pKa = 3.53EE68 pKa = 4.18YY69 pKa = 9.87TQAKK73 pKa = 8.41AVSMFNAGGYY83 pKa = 7.52STISDD88 pKa = 2.99ARR90 pKa = 11.84AAFVAAWTGDD100 pKa = 3.54VYY102 pKa = 11.72NHH104 pKa = 5.88YY105 pKa = 10.04LQYY108 pKa = 11.17GSQEE112 pKa = 4.38GINPSNAFDD121 pKa = 3.53EE122 pKa = 4.48SSYY125 pKa = 10.69YY126 pKa = 10.57ASKK129 pKa = 10.77LADD132 pKa = 3.66LQANADD138 pKa = 4.05TSAEE142 pKa = 3.99WATKK146 pKa = 10.28SVAEE150 pKa = 4.46LKK152 pKa = 10.99AFFNAQGFSALHH164 pKa = 6.47HH165 pKa = 5.94YY166 pKa = 9.72NVYY169 pKa = 10.4GAGEE173 pKa = 5.04GIPVTEE179 pKa = 4.39VPVSEE184 pKa = 4.45AVNSEE189 pKa = 3.57TDD191 pKa = 3.7EE192 pKa = 4.42IDD194 pKa = 3.4EE195 pKa = 4.32TDD197 pKa = 3.52EE198 pKa = 4.04TDD200 pKa = 3.39EE201 pKa = 4.84TGGTPAKK208 pKa = 10.47GMNFQLTTNDD218 pKa = 3.46DD219 pKa = 3.98SFTGGAANDD228 pKa = 4.12HH229 pKa = 6.87FYY231 pKa = 11.36GIYY234 pKa = 10.69DD235 pKa = 3.69GVTFLAQNTFNSGDD249 pKa = 3.62ILDD252 pKa = 4.59GAGGTDD258 pKa = 3.28TLHH261 pKa = 6.66INHH264 pKa = 6.42FEE266 pKa = 4.11DD267 pKa = 4.34VAIDD271 pKa = 4.32LPDD274 pKa = 3.89TLWTGISGIEE284 pKa = 4.21NIVMSTTGSGAHH296 pKa = 7.16DD297 pKa = 5.35IITGTNFNTAFASGGVDD314 pKa = 3.15LWTATTGAGAITIDD328 pKa = 3.48MASFTGTAKK337 pKa = 9.13VTADD341 pKa = 3.34SAAGAIVLSGVKK353 pKa = 10.31FSQIDD358 pKa = 3.56VTTTGSGAQTITSTGASDD376 pKa = 4.08VIVNATSSEE385 pKa = 4.1GATSITTGAGDD396 pKa = 3.69DD397 pKa = 4.38TIALHH402 pKa = 5.87ATSADD407 pKa = 3.28GLNTITAGAGADD419 pKa = 4.03TINLYY424 pKa = 11.39ANFASIDD431 pKa = 3.92KK432 pKa = 10.29IVQADD437 pKa = 4.1GDD439 pKa = 4.64SMASTANTITSNIATGDD456 pKa = 3.65TITFGNGLDD465 pKa = 3.13IVRR468 pKa = 11.84NFGVNDD474 pKa = 4.05NIDD477 pKa = 3.61VGTGGAAVSGLTMDD491 pKa = 4.39KK492 pKa = 11.14ADD494 pKa = 4.09FAATTTIFLSGTYY507 pKa = 10.43AEE509 pKa = 4.61DD510 pKa = 3.66TFTIAADD517 pKa = 3.66GGGTDD522 pKa = 3.99TLLLDD527 pKa = 3.81TTAVDD532 pKa = 3.87DD533 pKa = 4.27TDD535 pKa = 3.85IATADD540 pKa = 3.24TWILLSGVTSDD551 pKa = 4.2ALSAGSFVV559 pKa = 3.47
Molecular weight: 57.71 kDa
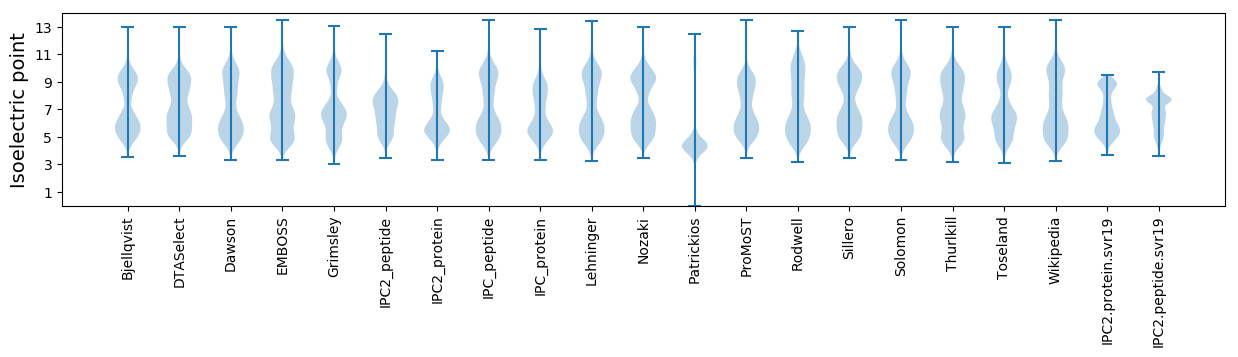
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S0FWG3|S0FWG3_9DELT 4HBT domain-containing protein OS=Desulfotignum phosphitoxidans DSM 13687 OX=1286635 GN=Dpo_6c02590 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 7.64RR12 pKa = 11.84TRR14 pKa = 11.84KK15 pKa = 9.77HH16 pKa = 5.74GFLKK20 pKa = 10.65RR21 pKa = 11.84MSTPGGKK28 pKa = 8.89RR29 pKa = 11.84VIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LALL44 pKa = 3.85
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 7.64RR12 pKa = 11.84TRR14 pKa = 11.84KK15 pKa = 9.77HH16 pKa = 5.74GFLKK20 pKa = 10.65RR21 pKa = 11.84MSTPGGKK28 pKa = 8.89RR29 pKa = 11.84VIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LALL44 pKa = 3.85
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1467878 |
29 |
3891 |
325.1 |
36.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.4 ± 0.04 | 1.367 ± 0.017 |
5.959 ± 0.031 | 5.648 ± 0.028 |
4.57 ± 0.028 | 7.201 ± 0.036 |
2.348 ± 0.016 | 6.937 ± 0.035 |
5.741 ± 0.033 | 9.696 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.972 ± 0.018 | 3.55 ± 0.025 |
4.624 ± 0.023 | 3.726 ± 0.024 |
5.375 ± 0.027 | 5.471 ± 0.021 |
5.671 ± 0.025 | 6.803 ± 0.03 |
1.112 ± 0.014 | 2.831 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
