
Microbacterium sp. Root61
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
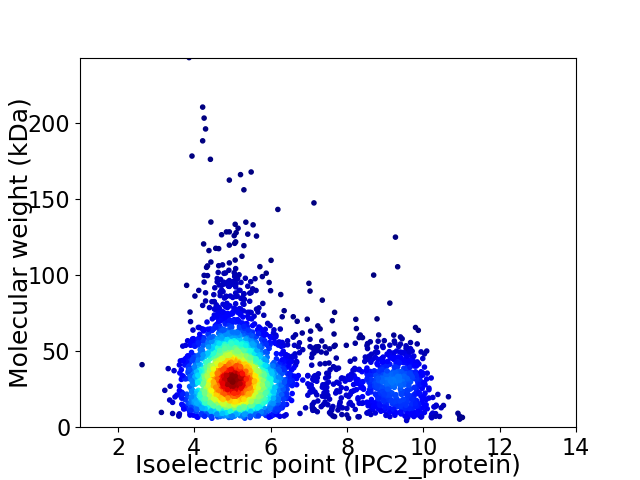
Virtual 2D-PAGE plot for 3629 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0T2L1A0|A0A0T2L1A0_9MICO Peptide synthetase OS=Microbacterium sp. Root61 OX=1736570 GN=ASD65_15575 PE=4 SV=1
MM1 pKa = 7.81AGTPTQNPDD10 pKa = 3.01ASWNISYY17 pKa = 8.93TITVKK22 pKa = 10.93NPGASPISATLTDD35 pKa = 4.9AFPATPAGWTLAGGVWNIAAQGGAPITNTTSAASPIWSGTLPANTTYY82 pKa = 10.6TYY84 pKa = 10.34IVSGKK89 pKa = 8.27LTPTAGATPIGDD101 pKa = 4.03CQTQGKK107 pKa = 9.19GLTNTATVTSGSVSDD122 pKa = 3.78TSGDD126 pKa = 3.74CVSIVTPPVTVTKK139 pKa = 10.46TDD141 pKa = 3.37GTVSQLVDD149 pKa = 3.66GTWQIDD155 pKa = 3.6YY156 pKa = 9.35TVTVTNGGTQATVYY170 pKa = 9.87TLTDD174 pKa = 3.55TPDD177 pKa = 3.24LGTGFTLVSGTWFGPAPVANTPIEE201 pKa = 4.39GGGFDD206 pKa = 3.2QYY208 pKa = 11.01VYY210 pKa = 10.87RR211 pKa = 11.84VIASFNPATPDD222 pKa = 3.88PEE224 pKa = 4.5LTCDD228 pKa = 3.49TTNGGAFFNKK238 pKa = 10.2ALVTFPGGTDD248 pKa = 3.7DD249 pKa = 5.27DD250 pKa = 4.67TGCGEE255 pKa = 4.25PEE257 pKa = 4.08SPTVEE262 pKa = 4.13KK263 pKa = 10.55SASAATQAPGGEE275 pKa = 3.9WTLTYY280 pKa = 10.31TVTVDD285 pKa = 3.44NTSEE289 pKa = 3.79MALAYY294 pKa = 7.62TASDD298 pKa = 3.66TPAALPAGVTLTTPWAVTGPAALNGGTATLTPGWTGTAPNTQFATGHH345 pKa = 6.41LLAGTAHH352 pKa = 7.27TYY354 pKa = 6.94TVKK357 pKa = 10.99AGVTLSAGVTPATLTCGQTPGGNGFWNSATVTNGVGTSDD396 pKa = 5.12DD397 pKa = 4.17SACVTVPFDD406 pKa = 4.12DD407 pKa = 3.76VGIVKK412 pKa = 7.88TTEE415 pKa = 4.19GVDD418 pKa = 3.84GPVEE422 pKa = 3.9SDD424 pKa = 3.59GVFKK428 pKa = 11.18YY429 pKa = 11.04VLTVTNHH436 pKa = 4.5GTRR439 pKa = 11.84AATNVKK445 pKa = 8.63VTDD448 pKa = 4.15PVPSRR453 pKa = 11.84LTVTGIDD460 pKa = 3.56LTDD463 pKa = 3.28ATGWTNDD470 pKa = 3.45NDD472 pKa = 4.05PDD474 pKa = 4.37FVGEE478 pKa = 4.31GNTVDD483 pKa = 3.34LTGPVSFGVGATAEE497 pKa = 3.98IVLTVKK503 pKa = 10.23VNPVPVPEE511 pKa = 4.4IPNLNEE517 pKa = 4.0GDD519 pKa = 3.93PVPTPEE525 pKa = 5.15LPMSTLVNEE534 pKa = 4.6ACVSADD540 pKa = 3.2MDD542 pKa = 4.34SVPGNDD548 pKa = 3.67CDD550 pKa = 4.34SVTVEE555 pKa = 4.54TKK557 pKa = 10.51DD558 pKa = 2.92IAAIVYY564 pKa = 7.03TRR566 pKa = 11.84CVGDD570 pKa = 3.47APLIGFVVAKK580 pKa = 9.1TPNLAALPVDD590 pKa = 4.72FTWTPNSPEE599 pKa = 4.58PDD601 pKa = 3.3TDD603 pKa = 3.6PAEE606 pKa = 4.27VAKK609 pKa = 10.37QYY611 pKa = 10.73PGGTATVSDD620 pKa = 3.6EE621 pKa = 4.24FAWVGTAFTPSGVSLDD637 pKa = 3.94YY638 pKa = 10.9PGWRR642 pKa = 11.84ALQASDD648 pKa = 3.65YY649 pKa = 10.64APGGGYY655 pKa = 9.69YY656 pKa = 9.75IPGTTDD662 pKa = 2.44VMTPTDD668 pKa = 4.06EE669 pKa = 4.83EE670 pKa = 4.09EE671 pKa = 4.22MIFNGLILDD680 pKa = 4.11PSEE683 pKa = 5.77LDD685 pKa = 3.61YY686 pKa = 11.45AWRR689 pKa = 11.84DD690 pKa = 3.44TTTVVLSVNPSMTFTVEE707 pKa = 4.17YY708 pKa = 10.16PDD710 pKa = 3.69ATPEE714 pKa = 4.18CFVARR719 pKa = 11.84HH720 pKa = 5.33TEE722 pKa = 3.92VQIEE726 pKa = 4.17KK727 pKa = 8.77TASVEE732 pKa = 3.9KK733 pKa = 9.81TDD735 pKa = 3.71PGKK738 pKa = 11.04SFTYY742 pKa = 9.86TLAAANVSDD751 pKa = 5.46DD752 pKa = 3.59SAADD756 pKa = 3.69GVVVTDD762 pKa = 5.32TIPADD767 pKa = 3.8LKK769 pKa = 10.17ITDD772 pKa = 3.85VSWTGKK778 pKa = 10.11GDD780 pKa = 4.0ANVFPNWSTCAVSGQNGAGYY800 pKa = 10.49GGTLTCEE807 pKa = 4.27LFGPLQPAGSGLGASAAPTITLSATVNASSKK838 pKa = 11.03ASVITNVGVVDD849 pKa = 4.38YY850 pKa = 8.91YY851 pKa = 11.43TFGDD855 pKa = 3.73PTDD858 pKa = 3.63TGRR861 pKa = 11.84DD862 pKa = 3.2ADD864 pKa = 4.12DD865 pKa = 4.14AVVLLSGLPATGGSALTPLILLGFLALLGGTATIVMIRR903 pKa = 11.84RR904 pKa = 11.84RR905 pKa = 11.84RR906 pKa = 11.84GSTKK910 pKa = 9.98PQLL913 pKa = 3.7
MM1 pKa = 7.81AGTPTQNPDD10 pKa = 3.01ASWNISYY17 pKa = 8.93TITVKK22 pKa = 10.93NPGASPISATLTDD35 pKa = 4.9AFPATPAGWTLAGGVWNIAAQGGAPITNTTSAASPIWSGTLPANTTYY82 pKa = 10.6TYY84 pKa = 10.34IVSGKK89 pKa = 8.27LTPTAGATPIGDD101 pKa = 4.03CQTQGKK107 pKa = 9.19GLTNTATVTSGSVSDD122 pKa = 3.78TSGDD126 pKa = 3.74CVSIVTPPVTVTKK139 pKa = 10.46TDD141 pKa = 3.37GTVSQLVDD149 pKa = 3.66GTWQIDD155 pKa = 3.6YY156 pKa = 9.35TVTVTNGGTQATVYY170 pKa = 9.87TLTDD174 pKa = 3.55TPDD177 pKa = 3.24LGTGFTLVSGTWFGPAPVANTPIEE201 pKa = 4.39GGGFDD206 pKa = 3.2QYY208 pKa = 11.01VYY210 pKa = 10.87RR211 pKa = 11.84VIASFNPATPDD222 pKa = 3.88PEE224 pKa = 4.5LTCDD228 pKa = 3.49TTNGGAFFNKK238 pKa = 10.2ALVTFPGGTDD248 pKa = 3.7DD249 pKa = 5.27DD250 pKa = 4.67TGCGEE255 pKa = 4.25PEE257 pKa = 4.08SPTVEE262 pKa = 4.13KK263 pKa = 10.55SASAATQAPGGEE275 pKa = 3.9WTLTYY280 pKa = 10.31TVTVDD285 pKa = 3.44NTSEE289 pKa = 3.79MALAYY294 pKa = 7.62TASDD298 pKa = 3.66TPAALPAGVTLTTPWAVTGPAALNGGTATLTPGWTGTAPNTQFATGHH345 pKa = 6.41LLAGTAHH352 pKa = 7.27TYY354 pKa = 6.94TVKK357 pKa = 10.99AGVTLSAGVTPATLTCGQTPGGNGFWNSATVTNGVGTSDD396 pKa = 5.12DD397 pKa = 4.17SACVTVPFDD406 pKa = 4.12DD407 pKa = 3.76VGIVKK412 pKa = 7.88TTEE415 pKa = 4.19GVDD418 pKa = 3.84GPVEE422 pKa = 3.9SDD424 pKa = 3.59GVFKK428 pKa = 11.18YY429 pKa = 11.04VLTVTNHH436 pKa = 4.5GTRR439 pKa = 11.84AATNVKK445 pKa = 8.63VTDD448 pKa = 4.15PVPSRR453 pKa = 11.84LTVTGIDD460 pKa = 3.56LTDD463 pKa = 3.28ATGWTNDD470 pKa = 3.45NDD472 pKa = 4.05PDD474 pKa = 4.37FVGEE478 pKa = 4.31GNTVDD483 pKa = 3.34LTGPVSFGVGATAEE497 pKa = 3.98IVLTVKK503 pKa = 10.23VNPVPVPEE511 pKa = 4.4IPNLNEE517 pKa = 4.0GDD519 pKa = 3.93PVPTPEE525 pKa = 5.15LPMSTLVNEE534 pKa = 4.6ACVSADD540 pKa = 3.2MDD542 pKa = 4.34SVPGNDD548 pKa = 3.67CDD550 pKa = 4.34SVTVEE555 pKa = 4.54TKK557 pKa = 10.51DD558 pKa = 2.92IAAIVYY564 pKa = 7.03TRR566 pKa = 11.84CVGDD570 pKa = 3.47APLIGFVVAKK580 pKa = 9.1TPNLAALPVDD590 pKa = 4.72FTWTPNSPEE599 pKa = 4.58PDD601 pKa = 3.3TDD603 pKa = 3.6PAEE606 pKa = 4.27VAKK609 pKa = 10.37QYY611 pKa = 10.73PGGTATVSDD620 pKa = 3.6EE621 pKa = 4.24FAWVGTAFTPSGVSLDD637 pKa = 3.94YY638 pKa = 10.9PGWRR642 pKa = 11.84ALQASDD648 pKa = 3.65YY649 pKa = 10.64APGGGYY655 pKa = 9.69YY656 pKa = 9.75IPGTTDD662 pKa = 2.44VMTPTDD668 pKa = 4.06EE669 pKa = 4.83EE670 pKa = 4.09EE671 pKa = 4.22MIFNGLILDD680 pKa = 4.11PSEE683 pKa = 5.77LDD685 pKa = 3.61YY686 pKa = 11.45AWRR689 pKa = 11.84DD690 pKa = 3.44TTTVVLSVNPSMTFTVEE707 pKa = 4.17YY708 pKa = 10.16PDD710 pKa = 3.69ATPEE714 pKa = 4.18CFVARR719 pKa = 11.84HH720 pKa = 5.33TEE722 pKa = 3.92VQIEE726 pKa = 4.17KK727 pKa = 8.77TASVEE732 pKa = 3.9KK733 pKa = 9.81TDD735 pKa = 3.71PGKK738 pKa = 11.04SFTYY742 pKa = 9.86TLAAANVSDD751 pKa = 5.46DD752 pKa = 3.59SAADD756 pKa = 3.69GVVVTDD762 pKa = 5.32TIPADD767 pKa = 3.8LKK769 pKa = 10.17ITDD772 pKa = 3.85VSWTGKK778 pKa = 10.11GDD780 pKa = 4.0ANVFPNWSTCAVSGQNGAGYY800 pKa = 10.49GGTLTCEE807 pKa = 4.27LFGPLQPAGSGLGASAAPTITLSATVNASSKK838 pKa = 11.03ASVITNVGVVDD849 pKa = 4.38YY850 pKa = 8.91YY851 pKa = 11.43TFGDD855 pKa = 3.73PTDD858 pKa = 3.63TGRR861 pKa = 11.84DD862 pKa = 3.2ADD864 pKa = 4.12DD865 pKa = 4.14AVVLLSGLPATGGSALTPLILLGFLALLGGTATIVMIRR903 pKa = 11.84RR904 pKa = 11.84RR905 pKa = 11.84RR906 pKa = 11.84GSTKK910 pKa = 9.98PQLL913 pKa = 3.7
Molecular weight: 93.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0T2KYN8|A0A0T2KYN8_9MICO HTH tetR-type domain-containing protein OS=Microbacterium sp. Root61 OX=1736570 GN=ASD65_10680 PE=4 SV=1
MM1 pKa = 7.57LGRR4 pKa = 11.84RR5 pKa = 11.84VGRR8 pKa = 11.84VGVIGAPVAKK18 pKa = 10.0AAVVGAAITPGPAPIAKK35 pKa = 9.67AAVVGAAVTPGRR47 pKa = 11.84SPVAKK52 pKa = 9.48AAVVGAVATPRR63 pKa = 11.84FRR65 pKa = 11.84RR66 pKa = 11.84II67 pKa = 3.12
MM1 pKa = 7.57LGRR4 pKa = 11.84RR5 pKa = 11.84VGRR8 pKa = 11.84VGVIGAPVAKK18 pKa = 10.0AAVVGAAITPGPAPIAKK35 pKa = 9.67AAVVGAAVTPGRR47 pKa = 11.84SPVAKK52 pKa = 9.48AAVVGAVATPRR63 pKa = 11.84FRR65 pKa = 11.84RR66 pKa = 11.84II67 pKa = 3.12
Molecular weight: 6.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1208867 |
38 |
2358 |
333.1 |
35.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.506 ± 0.062 | 0.507 ± 0.01 |
6.225 ± 0.035 | 5.533 ± 0.041 |
3.219 ± 0.024 | 8.944 ± 0.034 |
1.949 ± 0.022 | 4.893 ± 0.031 |
1.85 ± 0.028 | 10.079 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.919 ± 0.017 | 1.944 ± 0.024 |
5.412 ± 0.028 | 2.745 ± 0.021 |
6.978 ± 0.051 | 5.571 ± 0.029 |
6.185 ± 0.041 | 9.0 ± 0.042 |
1.549 ± 0.017 | 1.992 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
