
Streptomyces phage Wofford
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Karimacvirus; Streptomyces virus Wollford
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
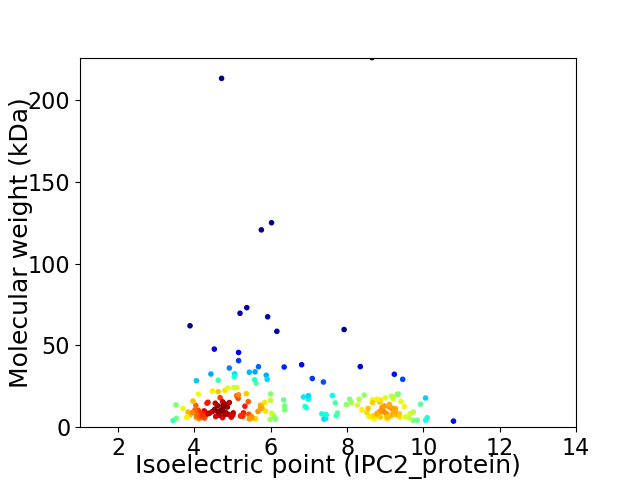
Virtual 2D-PAGE plot for 212 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
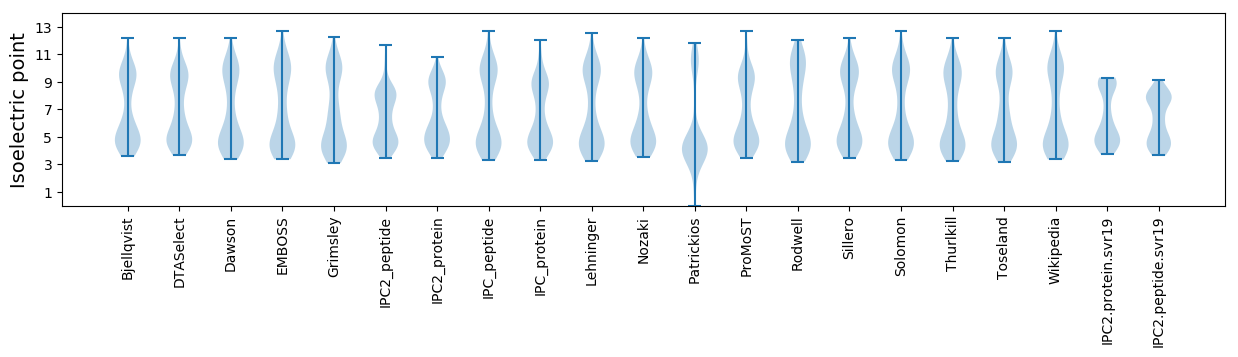
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345M9S3|A0A345M9S3_9CAUD Portal protein OS=Streptomyces phage Wofford OX=2283267 GN=48 PE=4 SV=1
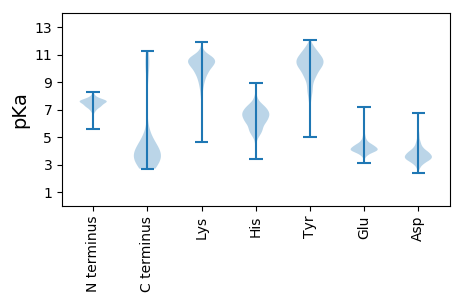
MM1 pKa = 7.76SYY3 pKa = 10.54QLQVLADD10 pKa = 3.64APFSYY15 pKa = 9.81WKK17 pKa = 10.48LDD19 pKa = 3.49GNGPVYY25 pKa = 10.33EE26 pKa = 4.92DD27 pKa = 3.18SAGSFRR33 pKa = 11.84QADD36 pKa = 3.94LTATAIPHH44 pKa = 6.43PALVTGSGNALVLQNSNRR62 pKa = 11.84LQMDD66 pKa = 3.38DD67 pKa = 3.45PVFNKK72 pKa = 10.35GYY74 pKa = 8.74EE75 pKa = 3.82ARR77 pKa = 11.84QWSLEE82 pKa = 3.77AWIKK86 pKa = 9.92PLSINGEE93 pKa = 3.91ISIMSHH99 pKa = 5.46SGNYY103 pKa = 9.83DD104 pKa = 3.14GLTITPTTINFRR116 pKa = 11.84TKK118 pKa = 10.81YY119 pKa = 8.41LTAPASEE126 pKa = 4.02INYY129 pKa = 9.55IYY131 pKa = 11.0NSTKK135 pKa = 9.48AFHH138 pKa = 6.13VVGVHH143 pKa = 5.12TNGKK147 pKa = 9.3NSLYY151 pKa = 10.82VDD153 pKa = 3.91GNLVAEE159 pKa = 4.45IDD161 pKa = 3.55ITDD164 pKa = 3.63EE165 pKa = 4.6QEE167 pKa = 3.12IDD169 pKa = 3.27AYY171 pKa = 11.5NFLTSDD177 pKa = 4.61LIAGEE182 pKa = 4.45SVSTSKK188 pKa = 10.64IAIDD192 pKa = 3.79SPAVYY197 pKa = 10.43AFALPADD204 pKa = 4.58TIKK207 pKa = 10.66KK208 pKa = 9.95HH209 pKa = 5.41YY210 pKa = 10.15DD211 pKa = 3.21CGVDD215 pKa = 3.67VEE217 pKa = 4.81SSDD220 pKa = 5.22AIAGFNDD227 pKa = 3.13ASHH230 pKa = 6.9WNFSDD235 pKa = 3.32EE236 pKa = 4.24SRR238 pKa = 11.84NIAVTKK244 pKa = 10.48LWDD247 pKa = 3.96TEE249 pKa = 4.38EE250 pKa = 4.06EE251 pKa = 4.36WPDD254 pKa = 3.53GVIDD258 pKa = 3.94NVVIANDD265 pKa = 4.6FIVPSYY271 pKa = 10.25EE272 pKa = 3.75QTEE275 pKa = 4.41TEE277 pKa = 4.25EE278 pKa = 4.33VVDD281 pKa = 4.12GLIVPIYY288 pKa = 9.53TNTSIPGTWIGSVPLGMTLAGISDD312 pKa = 4.67AIIRR316 pKa = 11.84YY317 pKa = 9.19RR318 pKa = 11.84GEE320 pKa = 3.83GSFTIEE326 pKa = 4.23CSTDD330 pKa = 4.26NGTTWQDD337 pKa = 2.47VDD339 pKa = 3.73NAPSFDD345 pKa = 4.33LDD347 pKa = 3.63EE348 pKa = 4.69TDD350 pKa = 3.86SIGIKK355 pKa = 10.4VSFTGGIVDD364 pKa = 3.84DD365 pKa = 4.15QSYY368 pKa = 10.62VDD370 pKa = 3.69YY371 pKa = 10.62VQVIGYY377 pKa = 7.66EE378 pKa = 4.12DD379 pKa = 3.97KK380 pKa = 11.15NINGTRR386 pKa = 11.84SDD388 pKa = 3.29RR389 pKa = 11.84AAVLNGTGISSIKK402 pKa = 8.94TYY404 pKa = 10.84QPIEE408 pKa = 4.0YY409 pKa = 10.25ADD411 pKa = 4.18DD412 pKa = 3.91NGLLILSGDD421 pKa = 3.17ISVGIDD427 pKa = 3.09SSYY430 pKa = 11.5DD431 pKa = 3.48GEE433 pKa = 4.47EE434 pKa = 4.11EE435 pKa = 5.35PGDD438 pKa = 3.97LDD440 pKa = 4.89INGIDD445 pKa = 3.52MWVKK449 pKa = 9.74PVAGNIISAGANSISRR465 pKa = 11.84VGNTITFSGYY475 pKa = 10.75SDD477 pKa = 3.37VTVNGVSVASGATVFVNDD495 pKa = 2.75SWYY498 pKa = 10.58HH499 pKa = 5.26IGGVFSTPGNYY510 pKa = 9.83PITIGTANARR520 pKa = 11.84IGPASAIYY528 pKa = 10.7GDD530 pKa = 3.73ISLAGLQQIYY540 pKa = 10.65ASYY543 pKa = 10.79LGLPGLTVDD552 pKa = 4.52DD553 pKa = 4.73EE554 pKa = 4.84SVIGITTPADD564 pKa = 3.35EE565 pKa = 4.84ALLYY569 pKa = 10.1AHH571 pKa = 6.72VWGITPAGG579 pKa = 3.48
MM1 pKa = 7.76SYY3 pKa = 10.54QLQVLADD10 pKa = 3.64APFSYY15 pKa = 9.81WKK17 pKa = 10.48LDD19 pKa = 3.49GNGPVYY25 pKa = 10.33EE26 pKa = 4.92DD27 pKa = 3.18SAGSFRR33 pKa = 11.84QADD36 pKa = 3.94LTATAIPHH44 pKa = 6.43PALVTGSGNALVLQNSNRR62 pKa = 11.84LQMDD66 pKa = 3.38DD67 pKa = 3.45PVFNKK72 pKa = 10.35GYY74 pKa = 8.74EE75 pKa = 3.82ARR77 pKa = 11.84QWSLEE82 pKa = 3.77AWIKK86 pKa = 9.92PLSINGEE93 pKa = 3.91ISIMSHH99 pKa = 5.46SGNYY103 pKa = 9.83DD104 pKa = 3.14GLTITPTTINFRR116 pKa = 11.84TKK118 pKa = 10.81YY119 pKa = 8.41LTAPASEE126 pKa = 4.02INYY129 pKa = 9.55IYY131 pKa = 11.0NSTKK135 pKa = 9.48AFHH138 pKa = 6.13VVGVHH143 pKa = 5.12TNGKK147 pKa = 9.3NSLYY151 pKa = 10.82VDD153 pKa = 3.91GNLVAEE159 pKa = 4.45IDD161 pKa = 3.55ITDD164 pKa = 3.63EE165 pKa = 4.6QEE167 pKa = 3.12IDD169 pKa = 3.27AYY171 pKa = 11.5NFLTSDD177 pKa = 4.61LIAGEE182 pKa = 4.45SVSTSKK188 pKa = 10.64IAIDD192 pKa = 3.79SPAVYY197 pKa = 10.43AFALPADD204 pKa = 4.58TIKK207 pKa = 10.66KK208 pKa = 9.95HH209 pKa = 5.41YY210 pKa = 10.15DD211 pKa = 3.21CGVDD215 pKa = 3.67VEE217 pKa = 4.81SSDD220 pKa = 5.22AIAGFNDD227 pKa = 3.13ASHH230 pKa = 6.9WNFSDD235 pKa = 3.32EE236 pKa = 4.24SRR238 pKa = 11.84NIAVTKK244 pKa = 10.48LWDD247 pKa = 3.96TEE249 pKa = 4.38EE250 pKa = 4.06EE251 pKa = 4.36WPDD254 pKa = 3.53GVIDD258 pKa = 3.94NVVIANDD265 pKa = 4.6FIVPSYY271 pKa = 10.25EE272 pKa = 3.75QTEE275 pKa = 4.41TEE277 pKa = 4.25EE278 pKa = 4.33VVDD281 pKa = 4.12GLIVPIYY288 pKa = 9.53TNTSIPGTWIGSVPLGMTLAGISDD312 pKa = 4.67AIIRR316 pKa = 11.84YY317 pKa = 9.19RR318 pKa = 11.84GEE320 pKa = 3.83GSFTIEE326 pKa = 4.23CSTDD330 pKa = 4.26NGTTWQDD337 pKa = 2.47VDD339 pKa = 3.73NAPSFDD345 pKa = 4.33LDD347 pKa = 3.63EE348 pKa = 4.69TDD350 pKa = 3.86SIGIKK355 pKa = 10.4VSFTGGIVDD364 pKa = 3.84DD365 pKa = 4.15QSYY368 pKa = 10.62VDD370 pKa = 3.69YY371 pKa = 10.62VQVIGYY377 pKa = 7.66EE378 pKa = 4.12DD379 pKa = 3.97KK380 pKa = 11.15NINGTRR386 pKa = 11.84SDD388 pKa = 3.29RR389 pKa = 11.84AAVLNGTGISSIKK402 pKa = 8.94TYY404 pKa = 10.84QPIEE408 pKa = 4.0YY409 pKa = 10.25ADD411 pKa = 4.18DD412 pKa = 3.91NGLLILSGDD421 pKa = 3.17ISVGIDD427 pKa = 3.09SSYY430 pKa = 11.5DD431 pKa = 3.48GEE433 pKa = 4.47EE434 pKa = 4.11EE435 pKa = 5.35PGDD438 pKa = 3.97LDD440 pKa = 4.89INGIDD445 pKa = 3.52MWVKK449 pKa = 9.74PVAGNIISAGANSISRR465 pKa = 11.84VGNTITFSGYY475 pKa = 10.75SDD477 pKa = 3.37VTVNGVSVASGATVFVNDD495 pKa = 2.75SWYY498 pKa = 10.58HH499 pKa = 5.26IGGVFSTPGNYY510 pKa = 9.83PITIGTANARR520 pKa = 11.84IGPASAIYY528 pKa = 10.7GDD530 pKa = 3.73ISLAGLQQIYY540 pKa = 10.65ASYY543 pKa = 10.79LGLPGLTVDD552 pKa = 4.52DD553 pKa = 4.73EE554 pKa = 4.84SVIGITTPADD564 pKa = 3.35EE565 pKa = 4.84ALLYY569 pKa = 10.1AHH571 pKa = 6.72VWGITPAGG579 pKa = 3.48
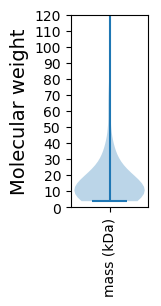
Molecular weight: 62.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345MA51|A0A345MA51_9CAUD Uncharacterized protein OS=Streptomyces phage Wofford OX=2283267 GN=236 PE=4 SV=1
MM1 pKa = 7.57SVVLDD6 pKa = 4.06YY7 pKa = 11.73NSVEE11 pKa = 4.11AFVTEE16 pKa = 4.08QRR18 pKa = 11.84KK19 pKa = 9.36LKK21 pKa = 10.82NDD23 pKa = 3.45VRR25 pKa = 11.84WDD27 pKa = 2.86GWTLVFFRR35 pKa = 11.84KK36 pKa = 9.47SRR38 pKa = 11.84NFSGWSKK45 pKa = 10.99PNGAYY50 pKa = 10.41RR51 pKa = 11.84NNNWGFEE58 pKa = 3.91TRR60 pKa = 11.84IAVGNDD66 pKa = 3.3GKK68 pKa = 10.3WRR70 pKa = 11.84VPSRR74 pKa = 11.84NVRR77 pKa = 11.84NSS79 pKa = 2.7
MM1 pKa = 7.57SVVLDD6 pKa = 4.06YY7 pKa = 11.73NSVEE11 pKa = 4.11AFVTEE16 pKa = 4.08QRR18 pKa = 11.84KK19 pKa = 9.36LKK21 pKa = 10.82NDD23 pKa = 3.45VRR25 pKa = 11.84WDD27 pKa = 2.86GWTLVFFRR35 pKa = 11.84KK36 pKa = 9.47SRR38 pKa = 11.84NFSGWSKK45 pKa = 10.99PNGAYY50 pKa = 10.41RR51 pKa = 11.84NNNWGFEE58 pKa = 3.91TRR60 pKa = 11.84IAVGNDD66 pKa = 3.3GKK68 pKa = 10.3WRR70 pKa = 11.84VPSRR74 pKa = 11.84NVRR77 pKa = 11.84NSS79 pKa = 2.7
Molecular weight: 9.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
33912 |
30 |
2084 |
160.0 |
17.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.212 ± 0.358 | 1.094 ± 0.131 |
6.585 ± 0.179 | 6.927 ± 0.321 |
3.786 ± 0.134 | 7.531 ± 0.26 |
1.743 ± 0.128 | 5.659 ± 0.165 |
6.487 ± 0.295 | 7.098 ± 0.182 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.978 ± 0.13 | 4.751 ± 0.141 |
3.651 ± 0.15 | 3.279 ± 0.179 |
5.547 ± 0.188 | 6.045 ± 0.253 |
5.939 ± 0.312 | 7.042 ± 0.203 |
1.881 ± 0.106 | 3.766 ± 0.151 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
