
Scyliorhinus torazame (Cloudy catshark)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Chondrichthyes; Elasmobranchii; Selachii; Galeomorphii; Galeoidea; Carcharhiniformes; Scyliorhinidae; Scyliorhinus
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
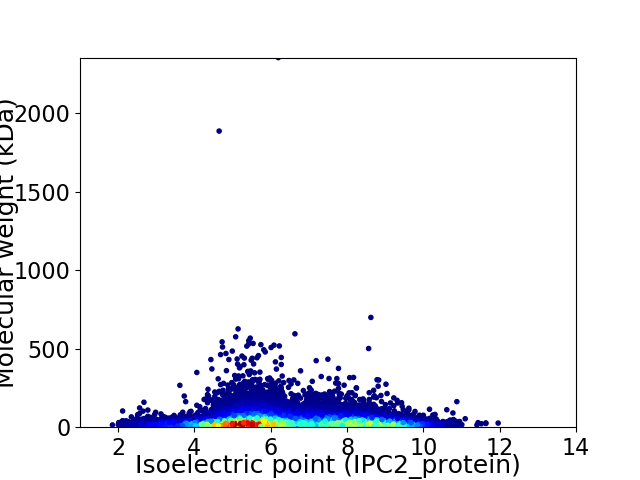
Virtual 2D-PAGE plot for 27602 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A401PT58|A0A401PT58_SCYTO Plus3 domain-containing protein OS=Scyliorhinus torazame OX=75743 GN=scyTo_0015464 PE=4 SV=1
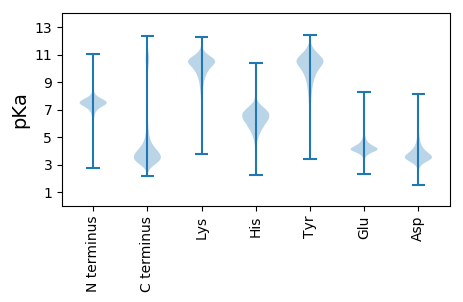
MM1 pKa = 7.07GTVFCWFDD9 pKa = 3.36GDD11 pKa = 4.19HH12 pKa = 5.92VLGMFHH18 pKa = 7.31GYY20 pKa = 9.54HH21 pKa = 5.39VLGMFDD27 pKa = 3.75GDD29 pKa = 3.79HH30 pKa = 6.0VLGVFDD36 pKa = 5.21GDD38 pKa = 3.93DD39 pKa = 3.43VLGVFDD45 pKa = 4.87GDD47 pKa = 3.79HH48 pKa = 5.87VLGVFDD54 pKa = 4.58GDD56 pKa = 3.53QVLGVFEE63 pKa = 4.86GDD65 pKa = 3.2HH66 pKa = 5.61VLGVFDD72 pKa = 4.72GDD74 pKa = 3.79HH75 pKa = 5.87VLGVFDD81 pKa = 4.72GDD83 pKa = 3.79HH84 pKa = 5.87VLGVFDD90 pKa = 4.6GDD92 pKa = 3.9HH93 pKa = 6.15VLGEE97 pKa = 4.12FDD99 pKa = 3.95GDD101 pKa = 4.19HH102 pKa = 6.52ILGVFDD108 pKa = 4.39GDD110 pKa = 3.7HH111 pKa = 5.69VLGVFEE117 pKa = 4.93GDD119 pKa = 3.2HH120 pKa = 5.61VLGVFDD126 pKa = 4.72GDD128 pKa = 3.79HH129 pKa = 5.87VLGVFDD135 pKa = 5.77GDD137 pKa = 3.56HH138 pKa = 6.95CLMGDD143 pKa = 3.47HH144 pKa = 6.33VLGVFDD150 pKa = 4.72GDD152 pKa = 4.0HH153 pKa = 5.92VLGVYY158 pKa = 10.57DD159 pKa = 4.04GDD161 pKa = 4.47HH162 pKa = 6.9ILRR165 pKa = 11.84VFDD168 pKa = 3.61GDD170 pKa = 3.62RR171 pKa = 11.84FLGVSDD177 pKa = 4.02GNHH180 pKa = 5.27VLGVFDD186 pKa = 4.09GDD188 pKa = 3.96YY189 pKa = 11.08VLGVFDD195 pKa = 4.73GNHH198 pKa = 5.33VLGVFDD204 pKa = 4.21GDD206 pKa = 3.37RR207 pKa = 11.84VLGVFDD213 pKa = 4.07WNHH216 pKa = 4.79VLRR219 pKa = 11.84VFDD222 pKa = 4.24GDD224 pKa = 3.77HH225 pKa = 5.82VLGVFDD231 pKa = 4.24GDD233 pKa = 3.29RR234 pKa = 11.84VLGVFHH240 pKa = 7.41GDD242 pKa = 3.01HH243 pKa = 6.09VLGVFDD249 pKa = 4.17GDD251 pKa = 3.96YY252 pKa = 11.08VLGVFDD258 pKa = 4.82GDD260 pKa = 3.79HH261 pKa = 5.87VLGVFDD267 pKa = 4.15GDD269 pKa = 3.97YY270 pKa = 10.43VLGMFDD276 pKa = 3.35VDD278 pKa = 3.49NVLGVFDD285 pKa = 4.12VDD287 pKa = 3.55HH288 pKa = 6.43VLGVFDD294 pKa = 4.24GDD296 pKa = 3.57RR297 pKa = 11.84VLGVYY302 pKa = 10.51DD303 pKa = 3.98GGHH306 pKa = 5.27VLGVFDD312 pKa = 3.96VDD314 pKa = 3.42RR315 pKa = 11.84VLGVFDD321 pKa = 4.79GDD323 pKa = 3.45HH324 pKa = 7.06DD325 pKa = 4.95LGVFDD330 pKa = 5.07GDD332 pKa = 3.73HH333 pKa = 5.87VLGVFDD339 pKa = 4.72GDD341 pKa = 3.79HH342 pKa = 5.87VLGVFDD348 pKa = 4.81GDD350 pKa = 3.94HH351 pKa = 5.92VLGGLMGTVFWEE363 pKa = 4.37CLMGTVFCWFDD374 pKa = 3.12GDD376 pKa = 4.24LVLGVFHH383 pKa = 7.34GDD385 pKa = 2.92HH386 pKa = 6.06VLGMFNGDD394 pKa = 3.05HH395 pKa = 5.97VLGVFDD401 pKa = 4.89GDD403 pKa = 4.35HH404 pKa = 6.41ILGVFDD410 pKa = 3.92GDD412 pKa = 3.47RR413 pKa = 11.84VLGVSDD419 pKa = 3.48RR420 pKa = 11.84TIVLWGPCAGWFDD433 pKa = 4.05GDD435 pKa = 3.87HH436 pKa = 5.9VLGVFDD442 pKa = 4.58GDD444 pKa = 3.63QVLGVFDD451 pKa = 4.94GDD453 pKa = 4.11DD454 pKa = 3.57VLGMFDD460 pKa = 3.86GDD462 pKa = 3.66HH463 pKa = 5.9VLGVFGGDD471 pKa = 2.92HH472 pKa = 5.83VLGVFDD478 pKa = 3.92GDD480 pKa = 4.18YY481 pKa = 11.11VHH483 pKa = 7.39GEE485 pKa = 3.75FDD487 pKa = 3.69GDD489 pKa = 3.81HH490 pKa = 5.99VLGVFDD496 pKa = 4.72GDD498 pKa = 3.79HH499 pKa = 5.87VLGVFDD505 pKa = 4.47GDD507 pKa = 3.87HH508 pKa = 5.7VVGVFDD514 pKa = 4.92GDD516 pKa = 3.99DD517 pKa = 3.48VLGMFDD523 pKa = 3.66GDD525 pKa = 3.36RR526 pKa = 11.84VLGVFDD532 pKa = 4.93GDD534 pKa = 5.17LILLVV539 pKa = 3.97
MM1 pKa = 7.07GTVFCWFDD9 pKa = 3.36GDD11 pKa = 4.19HH12 pKa = 5.92VLGMFHH18 pKa = 7.31GYY20 pKa = 9.54HH21 pKa = 5.39VLGMFDD27 pKa = 3.75GDD29 pKa = 3.79HH30 pKa = 6.0VLGVFDD36 pKa = 5.21GDD38 pKa = 3.93DD39 pKa = 3.43VLGVFDD45 pKa = 4.87GDD47 pKa = 3.79HH48 pKa = 5.87VLGVFDD54 pKa = 4.58GDD56 pKa = 3.53QVLGVFEE63 pKa = 4.86GDD65 pKa = 3.2HH66 pKa = 5.61VLGVFDD72 pKa = 4.72GDD74 pKa = 3.79HH75 pKa = 5.87VLGVFDD81 pKa = 4.72GDD83 pKa = 3.79HH84 pKa = 5.87VLGVFDD90 pKa = 4.6GDD92 pKa = 3.9HH93 pKa = 6.15VLGEE97 pKa = 4.12FDD99 pKa = 3.95GDD101 pKa = 4.19HH102 pKa = 6.52ILGVFDD108 pKa = 4.39GDD110 pKa = 3.7HH111 pKa = 5.69VLGVFEE117 pKa = 4.93GDD119 pKa = 3.2HH120 pKa = 5.61VLGVFDD126 pKa = 4.72GDD128 pKa = 3.79HH129 pKa = 5.87VLGVFDD135 pKa = 5.77GDD137 pKa = 3.56HH138 pKa = 6.95CLMGDD143 pKa = 3.47HH144 pKa = 6.33VLGVFDD150 pKa = 4.72GDD152 pKa = 4.0HH153 pKa = 5.92VLGVYY158 pKa = 10.57DD159 pKa = 4.04GDD161 pKa = 4.47HH162 pKa = 6.9ILRR165 pKa = 11.84VFDD168 pKa = 3.61GDD170 pKa = 3.62RR171 pKa = 11.84FLGVSDD177 pKa = 4.02GNHH180 pKa = 5.27VLGVFDD186 pKa = 4.09GDD188 pKa = 3.96YY189 pKa = 11.08VLGVFDD195 pKa = 4.73GNHH198 pKa = 5.33VLGVFDD204 pKa = 4.21GDD206 pKa = 3.37RR207 pKa = 11.84VLGVFDD213 pKa = 4.07WNHH216 pKa = 4.79VLRR219 pKa = 11.84VFDD222 pKa = 4.24GDD224 pKa = 3.77HH225 pKa = 5.82VLGVFDD231 pKa = 4.24GDD233 pKa = 3.29RR234 pKa = 11.84VLGVFHH240 pKa = 7.41GDD242 pKa = 3.01HH243 pKa = 6.09VLGVFDD249 pKa = 4.17GDD251 pKa = 3.96YY252 pKa = 11.08VLGVFDD258 pKa = 4.82GDD260 pKa = 3.79HH261 pKa = 5.87VLGVFDD267 pKa = 4.15GDD269 pKa = 3.97YY270 pKa = 10.43VLGMFDD276 pKa = 3.35VDD278 pKa = 3.49NVLGVFDD285 pKa = 4.12VDD287 pKa = 3.55HH288 pKa = 6.43VLGVFDD294 pKa = 4.24GDD296 pKa = 3.57RR297 pKa = 11.84VLGVYY302 pKa = 10.51DD303 pKa = 3.98GGHH306 pKa = 5.27VLGVFDD312 pKa = 3.96VDD314 pKa = 3.42RR315 pKa = 11.84VLGVFDD321 pKa = 4.79GDD323 pKa = 3.45HH324 pKa = 7.06DD325 pKa = 4.95LGVFDD330 pKa = 5.07GDD332 pKa = 3.73HH333 pKa = 5.87VLGVFDD339 pKa = 4.72GDD341 pKa = 3.79HH342 pKa = 5.87VLGVFDD348 pKa = 4.81GDD350 pKa = 3.94HH351 pKa = 5.92VLGGLMGTVFWEE363 pKa = 4.37CLMGTVFCWFDD374 pKa = 3.12GDD376 pKa = 4.24LVLGVFHH383 pKa = 7.34GDD385 pKa = 2.92HH386 pKa = 6.06VLGMFNGDD394 pKa = 3.05HH395 pKa = 5.97VLGVFDD401 pKa = 4.89GDD403 pKa = 4.35HH404 pKa = 6.41ILGVFDD410 pKa = 3.92GDD412 pKa = 3.47RR413 pKa = 11.84VLGVSDD419 pKa = 3.48RR420 pKa = 11.84TIVLWGPCAGWFDD433 pKa = 4.05GDD435 pKa = 3.87HH436 pKa = 5.9VLGVFDD442 pKa = 4.58GDD444 pKa = 3.63QVLGVFDD451 pKa = 4.94GDD453 pKa = 4.11DD454 pKa = 3.57VLGMFDD460 pKa = 3.86GDD462 pKa = 3.66HH463 pKa = 5.9VLGVFGGDD471 pKa = 2.92HH472 pKa = 5.83VLGVFDD478 pKa = 3.92GDD480 pKa = 4.18YY481 pKa = 11.11VHH483 pKa = 7.39GEE485 pKa = 3.75FDD487 pKa = 3.69GDD489 pKa = 3.81HH490 pKa = 5.99VLGVFDD496 pKa = 4.72GDD498 pKa = 3.79HH499 pKa = 5.87VLGVFDD505 pKa = 4.47GDD507 pKa = 3.87HH508 pKa = 5.7VVGVFDD514 pKa = 4.92GDD516 pKa = 3.99DD517 pKa = 3.48VLGMFDD523 pKa = 3.66GDD525 pKa = 3.36RR526 pKa = 11.84VLGVFDD532 pKa = 4.93GDD534 pKa = 5.17LILLVV539 pKa = 3.97
Molecular weight: 57.68 kDa
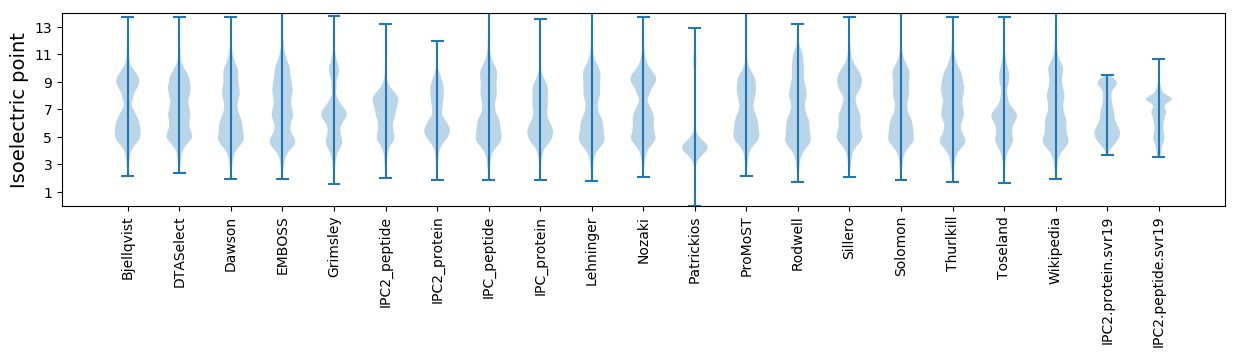
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A401Q8M1|A0A401Q8M1_SCYTO Uncharacterized protein (Fragment) OS=Scyliorhinus torazame OX=75743 GN=scyTo_0023172 PE=4 SV=1
MM1 pKa = 7.6SSGFRR6 pKa = 11.84LLPSGFRR13 pKa = 11.84LLPSRR18 pKa = 11.84FTVAIWIQTSAIRR31 pKa = 11.84IQTAVIRR38 pKa = 11.84IQTAVIRR45 pKa = 11.84IKK47 pKa = 10.01TAVTRR52 pKa = 11.84IQTAATRR59 pKa = 11.84IQSAAIRR66 pKa = 11.84IQTAVIRR73 pKa = 11.84IQTAVIRR80 pKa = 11.84IQIAVTRR87 pKa = 11.84IQTAATRR94 pKa = 11.84IQTVAIRR101 pKa = 11.84IQTVAIRR108 pKa = 11.84IQTAASWIQTAAIRR122 pKa = 11.84II123 pKa = 4.13
MM1 pKa = 7.6SSGFRR6 pKa = 11.84LLPSGFRR13 pKa = 11.84LLPSRR18 pKa = 11.84FTVAIWIQTSAIRR31 pKa = 11.84IQTAVIRR38 pKa = 11.84IQTAVIRR45 pKa = 11.84IKK47 pKa = 10.01TAVTRR52 pKa = 11.84IQTAATRR59 pKa = 11.84IQSAAIRR66 pKa = 11.84IQTAVIRR73 pKa = 11.84IQTAVIRR80 pKa = 11.84IQIAVTRR87 pKa = 11.84IQTAATRR94 pKa = 11.84IQTVAIRR101 pKa = 11.84IQTVAIRR108 pKa = 11.84IQTAASWIQTAAIRR122 pKa = 11.84II123 pKa = 4.13
Molecular weight: 13.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10337261 |
30 |
21134 |
374.5 |
42.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.419 ± 0.021 | 2.199 ± 0.014 |
5.405 ± 0.019 | 7.401 ± 0.026 |
3.544 ± 0.012 | 6.018 ± 0.026 |
2.482 ± 0.009 | 5.088 ± 0.017 |
6.092 ± 0.02 | 9.376 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.375 ± 0.009 | 4.347 ± 0.013 |
5.002 ± 0.019 | 4.781 ± 0.018 |
5.477 ± 0.021 | 8.066 ± 0.023 |
5.68 ± 0.018 | 6.316 ± 0.019 |
1.138 ± 0.007 | 2.794 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
