
Acidocella sp. MX-AZ02
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Acidocella; unclassified Acidocella
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
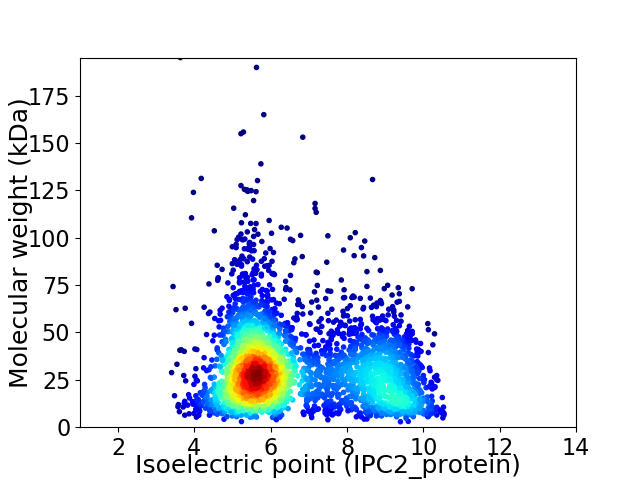
Virtual 2D-PAGE plot for 3506 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K5YLL3|K5YLL3_9PROT ImpA domain-containing protein OS=Acidocella sp. MX-AZ02 OX=1214225 GN=MXAZACID_08866 PE=4 SV=1
MM1 pKa = 7.57SGLTNAVNAALSGLQLFEE19 pKa = 5.71AGISTVSNNLANANTAGYY37 pKa = 7.74TVEE40 pKa = 4.36NVNAQTAQAAAGQPGAGVLPAQITRR65 pKa = 11.84AASGFAASQLRR76 pKa = 11.84TANSASAAASAQSTALTDD94 pKa = 3.48ISNALTNNGNVQTAINQFFSDD115 pKa = 3.82IGSLAASPSSSALRR129 pKa = 11.84QTVLSDD135 pKa = 2.95AGNVVGTFQTAATSIASVVSGQQAGLSSNVTAANQILQQLSTLNKK180 pKa = 10.06SLQTAPNSPSLLDD193 pKa = 3.67QRR195 pKa = 11.84DD196 pKa = 3.54SALNALSQYY205 pKa = 10.63LPVNIIPQSDD215 pKa = 3.71GGVVVATGGTVLVDD229 pKa = 3.21QSGVQALALAEE240 pKa = 4.1NANGQTTLTAGNNQMKK256 pKa = 8.92LTLGEE261 pKa = 4.21SDD263 pKa = 3.65GSLGATLANIVAGGQATQSLSALAAVFSAQVNTAQAQGLTPTGTSGTPIFSVPAPSVAPSGGNSGSAVITASLSNEE339 pKa = 3.61AALPANGGPFTLTYY353 pKa = 10.49NSTSGWSAVDD363 pKa = 3.32QANGQSYY370 pKa = 10.03IVGGTPPAFAGLALSISGTANNGDD394 pKa = 3.85SFTVNPAPGAANAIAVAATTPNDD417 pKa = 3.03IAAADD422 pKa = 4.5PYY424 pKa = 11.09VATNGTLQTDD434 pKa = 4.1GSVINKK440 pKa = 9.09NAGTINVGTDD450 pKa = 3.12SVTSSPASGSAVVPASYY467 pKa = 10.61YY468 pKa = 10.21GQNLLLTFTSASAYY482 pKa = 10.01NVSTLASPGTIIASGTLGSNGGNVAVAYY510 pKa = 9.2PSGLASGQYY519 pKa = 9.42WNLPLTGAPVAGDD532 pKa = 3.83TLSLSPGGSASGSNAQRR549 pKa = 11.84LANLWTATGTTSSGTLEE566 pKa = 3.84QSFVGLSTDD575 pKa = 3.81LGANAAAAQALATSTGSQVTAATTNLATIAGVNSDD610 pKa = 3.53QQAVLMTNYY619 pKa = 9.17EE620 pKa = 3.91QAYY623 pKa = 8.66QAAAKK628 pKa = 10.01AISAANTMFDD638 pKa = 3.68SLLNAII644 pKa = 4.92
MM1 pKa = 7.57SGLTNAVNAALSGLQLFEE19 pKa = 5.71AGISTVSNNLANANTAGYY37 pKa = 7.74TVEE40 pKa = 4.36NVNAQTAQAAAGQPGAGVLPAQITRR65 pKa = 11.84AASGFAASQLRR76 pKa = 11.84TANSASAAASAQSTALTDD94 pKa = 3.48ISNALTNNGNVQTAINQFFSDD115 pKa = 3.82IGSLAASPSSSALRR129 pKa = 11.84QTVLSDD135 pKa = 2.95AGNVVGTFQTAATSIASVVSGQQAGLSSNVTAANQILQQLSTLNKK180 pKa = 10.06SLQTAPNSPSLLDD193 pKa = 3.67QRR195 pKa = 11.84DD196 pKa = 3.54SALNALSQYY205 pKa = 10.63LPVNIIPQSDD215 pKa = 3.71GGVVVATGGTVLVDD229 pKa = 3.21QSGVQALALAEE240 pKa = 4.1NANGQTTLTAGNNQMKK256 pKa = 8.92LTLGEE261 pKa = 4.21SDD263 pKa = 3.65GSLGATLANIVAGGQATQSLSALAAVFSAQVNTAQAQGLTPTGTSGTPIFSVPAPSVAPSGGNSGSAVITASLSNEE339 pKa = 3.61AALPANGGPFTLTYY353 pKa = 10.49NSTSGWSAVDD363 pKa = 3.32QANGQSYY370 pKa = 10.03IVGGTPPAFAGLALSISGTANNGDD394 pKa = 3.85SFTVNPAPGAANAIAVAATTPNDD417 pKa = 3.03IAAADD422 pKa = 4.5PYY424 pKa = 11.09VATNGTLQTDD434 pKa = 4.1GSVINKK440 pKa = 9.09NAGTINVGTDD450 pKa = 3.12SVTSSPASGSAVVPASYY467 pKa = 10.61YY468 pKa = 10.21GQNLLLTFTSASAYY482 pKa = 10.01NVSTLASPGTIIASGTLGSNGGNVAVAYY510 pKa = 9.2PSGLASGQYY519 pKa = 9.42WNLPLTGAPVAGDD532 pKa = 3.83TLSLSPGGSASGSNAQRR549 pKa = 11.84LANLWTATGTTSSGTLEE566 pKa = 3.84QSFVGLSTDD575 pKa = 3.81LGANAAAAQALATSTGSQVTAATTNLATIAGVNSDD610 pKa = 3.53QQAVLMTNYY619 pKa = 9.17EE620 pKa = 3.91QAYY623 pKa = 8.66QAAAKK628 pKa = 10.01AISAANTMFDD638 pKa = 3.68SLLNAII644 pKa = 4.92
Molecular weight: 62.77 kDa
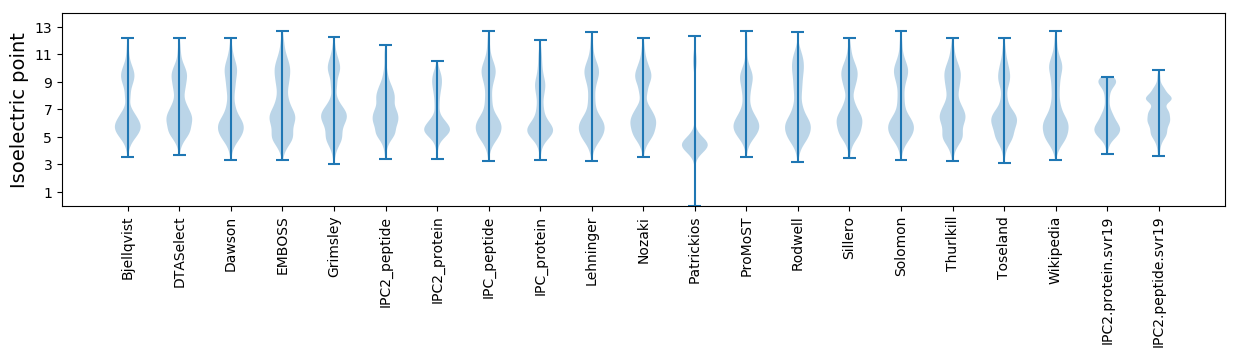
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K5ZEQ9|K5ZEQ9_9PROT TonB-dependent receptor OS=Acidocella sp. MX-AZ02 OX=1214225 GN=MXAZACID_17201 PE=4 SV=1
MM1 pKa = 7.69RR2 pKa = 11.84MEE4 pKa = 4.86ARR6 pKa = 11.84MHH8 pKa = 6.77DD9 pKa = 4.72LVTLGPWLRR18 pKa = 11.84RR19 pKa = 11.84FLSEE23 pKa = 4.97HH24 pKa = 6.12IVTEE28 pKa = 4.18RR29 pKa = 11.84NLARR33 pKa = 11.84NTRR36 pKa = 11.84SSYY39 pKa = 11.14RR40 pKa = 11.84DD41 pKa = 3.5TFSLLLPFISRR52 pKa = 11.84KK53 pKa = 8.65LRR55 pKa = 11.84KK56 pKa = 9.05PVDD59 pKa = 3.31RR60 pKa = 11.84LAVRR64 pKa = 11.84DD65 pKa = 4.24LSTQHH70 pKa = 6.18VLQFLAHH77 pKa = 6.31LEE79 pKa = 3.94EE80 pKa = 5.61DD81 pKa = 3.9RR82 pKa = 11.84GCSARR87 pKa = 11.84TRR89 pKa = 11.84NQRR92 pKa = 11.84LAAIRR97 pKa = 11.84AFARR101 pKa = 11.84FIGSRR106 pKa = 11.84DD107 pKa = 3.33PAHH110 pKa = 7.05IEE112 pKa = 3.65WCGHH116 pKa = 4.01IRR118 pKa = 11.84AIASKK123 pKa = 10.76KK124 pKa = 10.8SMQQPVGWLARR135 pKa = 11.84PEE137 pKa = 4.09MEE139 pKa = 5.49AMLAVPDD146 pKa = 3.82RR147 pKa = 11.84KK148 pKa = 8.52TPRR151 pKa = 11.84GRR153 pKa = 11.84DD154 pKa = 3.33EE155 pKa = 4.0YY156 pKa = 11.8ALLLFLYY163 pKa = 8.54NTGARR168 pKa = 11.84VSEE171 pKa = 3.95ATQLKK176 pKa = 9.99VRR178 pKa = 11.84DD179 pKa = 3.78LQLEE183 pKa = 4.21HH184 pKa = 7.04GNRR187 pKa = 11.84GHH189 pKa = 7.58DD190 pKa = 3.59LVTLHH195 pKa = 6.75GKK197 pKa = 9.45GGKK200 pKa = 7.38TRR202 pKa = 11.84QCPLWPEE209 pKa = 4.43TKK211 pKa = 10.29AVLAQRR217 pKa = 11.84VLGRR221 pKa = 11.84AAEE224 pKa = 4.04NAVFVSRR231 pKa = 11.84LGTPFTRR238 pKa = 11.84FGVYY242 pKa = 10.12RR243 pKa = 11.84LIEE246 pKa = 4.05RR247 pKa = 11.84CAARR251 pKa = 11.84VPALAGRR258 pKa = 11.84TITPHH263 pKa = 6.44VIRR266 pKa = 11.84HH267 pKa = 4.71TTACHH272 pKa = 5.6LVLAGVDD279 pKa = 3.5INTVRR284 pKa = 11.84AWLGHH289 pKa = 4.67VSISTTNIYY298 pKa = 11.05AEE300 pKa = 3.93IDD302 pKa = 3.25ITLKK306 pKa = 10.45AQAVALCEE314 pKa = 4.01VGQPQLGRR322 pKa = 11.84SWKK325 pKa = 9.71NDD327 pKa = 3.16KK328 pKa = 11.22DD329 pKa = 3.59LMAFLKK335 pKa = 10.89SLL337 pKa = 3.63
MM1 pKa = 7.69RR2 pKa = 11.84MEE4 pKa = 4.86ARR6 pKa = 11.84MHH8 pKa = 6.77DD9 pKa = 4.72LVTLGPWLRR18 pKa = 11.84RR19 pKa = 11.84FLSEE23 pKa = 4.97HH24 pKa = 6.12IVTEE28 pKa = 4.18RR29 pKa = 11.84NLARR33 pKa = 11.84NTRR36 pKa = 11.84SSYY39 pKa = 11.14RR40 pKa = 11.84DD41 pKa = 3.5TFSLLLPFISRR52 pKa = 11.84KK53 pKa = 8.65LRR55 pKa = 11.84KK56 pKa = 9.05PVDD59 pKa = 3.31RR60 pKa = 11.84LAVRR64 pKa = 11.84DD65 pKa = 4.24LSTQHH70 pKa = 6.18VLQFLAHH77 pKa = 6.31LEE79 pKa = 3.94EE80 pKa = 5.61DD81 pKa = 3.9RR82 pKa = 11.84GCSARR87 pKa = 11.84TRR89 pKa = 11.84NQRR92 pKa = 11.84LAAIRR97 pKa = 11.84AFARR101 pKa = 11.84FIGSRR106 pKa = 11.84DD107 pKa = 3.33PAHH110 pKa = 7.05IEE112 pKa = 3.65WCGHH116 pKa = 4.01IRR118 pKa = 11.84AIASKK123 pKa = 10.76KK124 pKa = 10.8SMQQPVGWLARR135 pKa = 11.84PEE137 pKa = 4.09MEE139 pKa = 5.49AMLAVPDD146 pKa = 3.82RR147 pKa = 11.84KK148 pKa = 8.52TPRR151 pKa = 11.84GRR153 pKa = 11.84DD154 pKa = 3.33EE155 pKa = 4.0YY156 pKa = 11.8ALLLFLYY163 pKa = 8.54NTGARR168 pKa = 11.84VSEE171 pKa = 3.95ATQLKK176 pKa = 9.99VRR178 pKa = 11.84DD179 pKa = 3.78LQLEE183 pKa = 4.21HH184 pKa = 7.04GNRR187 pKa = 11.84GHH189 pKa = 7.58DD190 pKa = 3.59LVTLHH195 pKa = 6.75GKK197 pKa = 9.45GGKK200 pKa = 7.38TRR202 pKa = 11.84QCPLWPEE209 pKa = 4.43TKK211 pKa = 10.29AVLAQRR217 pKa = 11.84VLGRR221 pKa = 11.84AAEE224 pKa = 4.04NAVFVSRR231 pKa = 11.84LGTPFTRR238 pKa = 11.84FGVYY242 pKa = 10.12RR243 pKa = 11.84LIEE246 pKa = 4.05RR247 pKa = 11.84CAARR251 pKa = 11.84VPALAGRR258 pKa = 11.84TITPHH263 pKa = 6.44VIRR266 pKa = 11.84HH267 pKa = 4.71TTACHH272 pKa = 5.6LVLAGVDD279 pKa = 3.5INTVRR284 pKa = 11.84AWLGHH289 pKa = 4.67VSISTTNIYY298 pKa = 11.05AEE300 pKa = 3.93IDD302 pKa = 3.25ITLKK306 pKa = 10.45AQAVALCEE314 pKa = 4.01VGQPQLGRR322 pKa = 11.84SWKK325 pKa = 9.71NDD327 pKa = 3.16KK328 pKa = 11.22DD329 pKa = 3.59LMAFLKK335 pKa = 10.89SLL337 pKa = 3.63
Molecular weight: 38.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1058590 |
25 |
1979 |
301.9 |
32.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.943 ± 0.06 | 0.9 ± 0.013 |
4.836 ± 0.031 | 5.244 ± 0.044 |
3.658 ± 0.026 | 8.871 ± 0.05 |
2.111 ± 0.019 | 5.028 ± 0.031 |
3.103 ± 0.033 | 11.161 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.425 ± 0.019 | 2.652 ± 0.033 |
5.503 ± 0.04 | 3.678 ± 0.03 |
6.432 ± 0.046 | 5.1 ± 0.04 |
5.058 ± 0.037 | 6.772 ± 0.033 |
1.326 ± 0.018 | 2.199 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
