
Simian foamy virus type 3 (strain LK3) (SFVagm) (SFV-3)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Spumavirus; African green monkey simian foamy virus; Simian foamy virus 3
Average proteome isoelectric point is 7.6
Get precalculated fractions of proteins
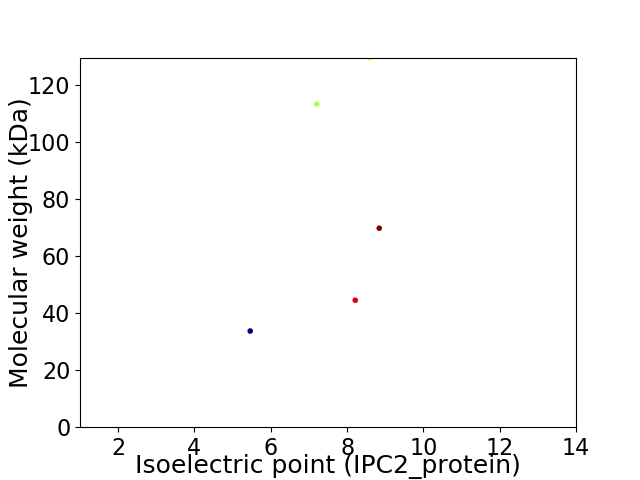
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P27403|BEL2_SFV3L Protein Bel-2 OS=Simian foamy virus type 3 (strain LK3) OX=11644 GN=bel2 PE=3 SV=2
MM1 pKa = 7.71ASWEE5 pKa = 4.21KK6 pKa = 10.64EE7 pKa = 3.71KK8 pKa = 11.0EE9 pKa = 4.02LAHH12 pKa = 6.2LHH14 pKa = 5.49QPEE17 pKa = 5.23DD18 pKa = 4.06DD19 pKa = 4.39PLPDD23 pKa = 4.64LSLLLDD29 pKa = 3.73MDD31 pKa = 3.69QFEE34 pKa = 4.58PTEE37 pKa = 4.73GPDD40 pKa = 3.64SNPGAEE46 pKa = 4.66KK47 pKa = 9.96IYY49 pKa = 10.63LQLQVAPGDD58 pKa = 3.95PSEE61 pKa = 4.07KK62 pKa = 9.32TYY64 pKa = 11.27KK65 pKa = 10.31FGYY68 pKa = 8.71EE69 pKa = 4.03DD70 pKa = 5.55KK71 pKa = 10.72EE72 pKa = 4.31AQNPDD77 pKa = 3.03LKK79 pKa = 10.36MRR81 pKa = 11.84NWVPDD86 pKa = 3.93PEE88 pKa = 5.25KK89 pKa = 9.98MSKK92 pKa = 8.43WACARR97 pKa = 11.84LILCGLYY104 pKa = 9.7NAKK107 pKa = 9.78KK108 pKa = 10.38AKK110 pKa = 9.86EE111 pKa = 4.17LLKK114 pKa = 9.77MDD116 pKa = 4.57YY117 pKa = 10.23DD118 pKa = 3.92IHH120 pKa = 6.37WEE122 pKa = 3.86QSKK125 pKa = 10.86EE126 pKa = 3.66DD127 pKa = 3.54SQYY130 pKa = 11.16FEE132 pKa = 5.01IEE134 pKa = 4.29YY135 pKa = 9.15HH136 pKa = 6.97CKK138 pKa = 8.74MCMTVIHH145 pKa = 6.6EE146 pKa = 4.43PMPVSYY152 pKa = 10.56DD153 pKa = 3.16KK154 pKa = 10.82KK155 pKa = 7.69TGLWIKK161 pKa = 9.43MGPLRR166 pKa = 11.84GDD168 pKa = 2.98IGSVVHH174 pKa = 5.51TCRR177 pKa = 11.84RR178 pKa = 11.84HH179 pKa = 4.64YY180 pKa = 9.08EE181 pKa = 3.92RR182 pKa = 11.84CMSALPSSGEE192 pKa = 3.7PLKK195 pKa = 10.87PRR197 pKa = 11.84VRR199 pKa = 11.84ANPVRR204 pKa = 11.84RR205 pKa = 11.84YY206 pKa = 9.68RR207 pKa = 11.84EE208 pKa = 3.49KK209 pKa = 11.04SLIVADD215 pKa = 4.19RR216 pKa = 11.84PKK218 pKa = 10.16RR219 pKa = 11.84SRR221 pKa = 11.84WGVAPRR227 pKa = 11.84EE228 pKa = 4.08QPNTSSGDD236 pKa = 3.34AMALMPGPCGPFNMDD251 pKa = 3.55PPGCLLEE258 pKa = 4.29RR259 pKa = 11.84VPGSEE264 pKa = 4.13PGTSEE269 pKa = 3.62MALAMSGGPFWEE281 pKa = 3.89QVYY284 pKa = 10.04RR285 pKa = 11.84DD286 pKa = 4.56SISGPPTGPSEE297 pKa = 4.06NN298 pKa = 3.71
MM1 pKa = 7.71ASWEE5 pKa = 4.21KK6 pKa = 10.64EE7 pKa = 3.71KK8 pKa = 11.0EE9 pKa = 4.02LAHH12 pKa = 6.2LHH14 pKa = 5.49QPEE17 pKa = 5.23DD18 pKa = 4.06DD19 pKa = 4.39PLPDD23 pKa = 4.64LSLLLDD29 pKa = 3.73MDD31 pKa = 3.69QFEE34 pKa = 4.58PTEE37 pKa = 4.73GPDD40 pKa = 3.64SNPGAEE46 pKa = 4.66KK47 pKa = 9.96IYY49 pKa = 10.63LQLQVAPGDD58 pKa = 3.95PSEE61 pKa = 4.07KK62 pKa = 9.32TYY64 pKa = 11.27KK65 pKa = 10.31FGYY68 pKa = 8.71EE69 pKa = 4.03DD70 pKa = 5.55KK71 pKa = 10.72EE72 pKa = 4.31AQNPDD77 pKa = 3.03LKK79 pKa = 10.36MRR81 pKa = 11.84NWVPDD86 pKa = 3.93PEE88 pKa = 5.25KK89 pKa = 9.98MSKK92 pKa = 8.43WACARR97 pKa = 11.84LILCGLYY104 pKa = 9.7NAKK107 pKa = 9.78KK108 pKa = 10.38AKK110 pKa = 9.86EE111 pKa = 4.17LLKK114 pKa = 9.77MDD116 pKa = 4.57YY117 pKa = 10.23DD118 pKa = 3.92IHH120 pKa = 6.37WEE122 pKa = 3.86QSKK125 pKa = 10.86EE126 pKa = 3.66DD127 pKa = 3.54SQYY130 pKa = 11.16FEE132 pKa = 5.01IEE134 pKa = 4.29YY135 pKa = 9.15HH136 pKa = 6.97CKK138 pKa = 8.74MCMTVIHH145 pKa = 6.6EE146 pKa = 4.43PMPVSYY152 pKa = 10.56DD153 pKa = 3.16KK154 pKa = 10.82KK155 pKa = 7.69TGLWIKK161 pKa = 9.43MGPLRR166 pKa = 11.84GDD168 pKa = 2.98IGSVVHH174 pKa = 5.51TCRR177 pKa = 11.84RR178 pKa = 11.84HH179 pKa = 4.64YY180 pKa = 9.08EE181 pKa = 3.92RR182 pKa = 11.84CMSALPSSGEE192 pKa = 3.7PLKK195 pKa = 10.87PRR197 pKa = 11.84VRR199 pKa = 11.84ANPVRR204 pKa = 11.84RR205 pKa = 11.84YY206 pKa = 9.68RR207 pKa = 11.84EE208 pKa = 3.49KK209 pKa = 11.04SLIVADD215 pKa = 4.19RR216 pKa = 11.84PKK218 pKa = 10.16RR219 pKa = 11.84SRR221 pKa = 11.84WGVAPRR227 pKa = 11.84EE228 pKa = 4.08QPNTSSGDD236 pKa = 3.34AMALMPGPCGPFNMDD251 pKa = 3.55PPGCLLEE258 pKa = 4.29RR259 pKa = 11.84VPGSEE264 pKa = 4.13PGTSEE269 pKa = 3.62MALAMSGGPFWEE281 pKa = 3.89QVYY284 pKa = 10.04RR285 pKa = 11.84DD286 pKa = 4.56SISGPPTGPSEE297 pKa = 4.06NN298 pKa = 3.71
Molecular weight: 33.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P27401|POL_SFV3L Pro-Pol polyprotein OS=Simian foamy virus type 3 (strain LK3) OX=11644 GN=pol PE=3 SV=2
MM1 pKa = 7.91GDD3 pKa = 3.22HH4 pKa = 6.6NLNVQEE10 pKa = 5.05LLNLFQNLGIPRR22 pKa = 11.84QPNHH26 pKa = 6.81RR27 pKa = 11.84EE28 pKa = 4.07VIGLRR33 pKa = 11.84MLGGWWGPGTRR44 pKa = 11.84YY45 pKa = 10.03ILVSIFLQDD54 pKa = 4.79DD55 pKa = 3.54SGQPLQQPRR64 pKa = 11.84WRR66 pKa = 11.84PEE68 pKa = 3.58GRR70 pKa = 11.84PVNPLVHH77 pKa = 5.94NTIEE81 pKa = 4.36APWGEE86 pKa = 3.9LRR88 pKa = 11.84QAFEE92 pKa = 4.85DD93 pKa = 3.92LDD95 pKa = 3.94VAEE98 pKa = 4.6GTLRR102 pKa = 11.84FGPLANGNWIPGDD115 pKa = 3.8EE116 pKa = 4.31YY117 pKa = 11.66SMEE120 pKa = 4.14FQPPLAQEE128 pKa = 4.24IAQMQRR134 pKa = 11.84DD135 pKa = 4.24EE136 pKa = 4.88LEE138 pKa = 4.5EE139 pKa = 4.36ILDD142 pKa = 3.6ITGQICAQVIDD153 pKa = 4.62LVDD156 pKa = 3.47MQDD159 pKa = 3.02AQIRR163 pKa = 11.84GLEE166 pKa = 3.83RR167 pKa = 11.84RR168 pKa = 11.84IQDD171 pKa = 3.49RR172 pKa = 11.84LGLRR176 pKa = 11.84DD177 pKa = 4.04NLPVAGIQAPPSSPIGQPIASSSLQPIPGSSSSPADD213 pKa = 4.0LDD215 pKa = 5.34GIWTPRR221 pKa = 11.84QIDD224 pKa = 3.5PRR226 pKa = 11.84LSRR229 pKa = 11.84VAYY232 pKa = 10.53NPFLPGSSDD241 pKa = 3.2GSGGSIPVQPSAPPAVLPSLPSLPAPVSQPIIQYY275 pKa = 8.61VAQPPVPAPQAIPIQHH291 pKa = 5.9IRR293 pKa = 11.84AVTGNTPTNPRR304 pKa = 11.84DD305 pKa = 3.23IPMWLGRR312 pKa = 11.84HH313 pKa = 4.88SAAIEE318 pKa = 4.42GVFPMTTPDD327 pKa = 3.19LRR329 pKa = 11.84CRR331 pKa = 11.84VVNALIGGSLGLSLEE346 pKa = 5.09PIHH349 pKa = 6.19CVNWAAVVAALYY361 pKa = 10.62VRR363 pKa = 11.84THH365 pKa = 5.82GSYY368 pKa = 10.49PIHH371 pKa = 6.65EE372 pKa = 4.66LANVLRR378 pKa = 11.84AVVTQEE384 pKa = 3.63GVATGFQLGIMLSNQDD400 pKa = 3.81YY401 pKa = 10.0NLVWGILRR409 pKa = 11.84PLLPGQAVVTAMQQRR424 pKa = 11.84LDD426 pKa = 3.73QEE428 pKa = 4.49VNDD431 pKa = 4.23AARR434 pKa = 11.84ITSFNGHH441 pKa = 6.94LNDD444 pKa = 3.95IYY446 pKa = 11.4QLLGLNARR454 pKa = 11.84GQSIARR460 pKa = 11.84AQSASTSGNSASAGRR475 pKa = 11.84GRR477 pKa = 11.84RR478 pKa = 11.84GQRR481 pKa = 11.84TQQQAGRR488 pKa = 11.84QQQQQTRR495 pKa = 11.84RR496 pKa = 11.84TNQGNQGQRR505 pKa = 11.84DD506 pKa = 3.67NNQRR510 pKa = 11.84QSSGGNQGQRR520 pKa = 11.84GQGGYY525 pKa = 9.34DD526 pKa = 3.32LRR528 pKa = 11.84PRR530 pKa = 11.84TYY532 pKa = 9.72QPQRR536 pKa = 11.84YY537 pKa = 8.66GGGRR541 pKa = 11.84GRR543 pKa = 11.84RR544 pKa = 11.84WNDD547 pKa = 2.87NQQQQQAQPGRR558 pKa = 11.84SSDD561 pKa = 3.6QPRR564 pKa = 11.84SQSQQPQPEE573 pKa = 3.9ARR575 pKa = 11.84GDD577 pKa = 3.46QSRR580 pKa = 11.84TSGAGRR586 pKa = 11.84GQQGRR591 pKa = 11.84GNQNRR596 pKa = 11.84NQRR599 pKa = 11.84RR600 pKa = 11.84ADD602 pKa = 3.49ANNTRR607 pKa = 11.84NVDD610 pKa = 3.53TVTATTTSSSTASSGQNGSSTTPPASGSRR639 pKa = 11.84NQGDD643 pKa = 3.19
MM1 pKa = 7.91GDD3 pKa = 3.22HH4 pKa = 6.6NLNVQEE10 pKa = 5.05LLNLFQNLGIPRR22 pKa = 11.84QPNHH26 pKa = 6.81RR27 pKa = 11.84EE28 pKa = 4.07VIGLRR33 pKa = 11.84MLGGWWGPGTRR44 pKa = 11.84YY45 pKa = 10.03ILVSIFLQDD54 pKa = 4.79DD55 pKa = 3.54SGQPLQQPRR64 pKa = 11.84WRR66 pKa = 11.84PEE68 pKa = 3.58GRR70 pKa = 11.84PVNPLVHH77 pKa = 5.94NTIEE81 pKa = 4.36APWGEE86 pKa = 3.9LRR88 pKa = 11.84QAFEE92 pKa = 4.85DD93 pKa = 3.92LDD95 pKa = 3.94VAEE98 pKa = 4.6GTLRR102 pKa = 11.84FGPLANGNWIPGDD115 pKa = 3.8EE116 pKa = 4.31YY117 pKa = 11.66SMEE120 pKa = 4.14FQPPLAQEE128 pKa = 4.24IAQMQRR134 pKa = 11.84DD135 pKa = 4.24EE136 pKa = 4.88LEE138 pKa = 4.5EE139 pKa = 4.36ILDD142 pKa = 3.6ITGQICAQVIDD153 pKa = 4.62LVDD156 pKa = 3.47MQDD159 pKa = 3.02AQIRR163 pKa = 11.84GLEE166 pKa = 3.83RR167 pKa = 11.84RR168 pKa = 11.84IQDD171 pKa = 3.49RR172 pKa = 11.84LGLRR176 pKa = 11.84DD177 pKa = 4.04NLPVAGIQAPPSSPIGQPIASSSLQPIPGSSSSPADD213 pKa = 4.0LDD215 pKa = 5.34GIWTPRR221 pKa = 11.84QIDD224 pKa = 3.5PRR226 pKa = 11.84LSRR229 pKa = 11.84VAYY232 pKa = 10.53NPFLPGSSDD241 pKa = 3.2GSGGSIPVQPSAPPAVLPSLPSLPAPVSQPIIQYY275 pKa = 8.61VAQPPVPAPQAIPIQHH291 pKa = 5.9IRR293 pKa = 11.84AVTGNTPTNPRR304 pKa = 11.84DD305 pKa = 3.23IPMWLGRR312 pKa = 11.84HH313 pKa = 4.88SAAIEE318 pKa = 4.42GVFPMTTPDD327 pKa = 3.19LRR329 pKa = 11.84CRR331 pKa = 11.84VVNALIGGSLGLSLEE346 pKa = 5.09PIHH349 pKa = 6.19CVNWAAVVAALYY361 pKa = 10.62VRR363 pKa = 11.84THH365 pKa = 5.82GSYY368 pKa = 10.49PIHH371 pKa = 6.65EE372 pKa = 4.66LANVLRR378 pKa = 11.84AVVTQEE384 pKa = 3.63GVATGFQLGIMLSNQDD400 pKa = 3.81YY401 pKa = 10.0NLVWGILRR409 pKa = 11.84PLLPGQAVVTAMQQRR424 pKa = 11.84LDD426 pKa = 3.73QEE428 pKa = 4.49VNDD431 pKa = 4.23AARR434 pKa = 11.84ITSFNGHH441 pKa = 6.94LNDD444 pKa = 3.95IYY446 pKa = 11.4QLLGLNARR454 pKa = 11.84GQSIARR460 pKa = 11.84AQSASTSGNSASAGRR475 pKa = 11.84GRR477 pKa = 11.84RR478 pKa = 11.84GQRR481 pKa = 11.84TQQQAGRR488 pKa = 11.84QQQQQTRR495 pKa = 11.84RR496 pKa = 11.84TNQGNQGQRR505 pKa = 11.84DD506 pKa = 3.67NNQRR510 pKa = 11.84QSSGGNQGQRR520 pKa = 11.84GQGGYY525 pKa = 9.34DD526 pKa = 3.32LRR528 pKa = 11.84PRR530 pKa = 11.84TYY532 pKa = 9.72QPQRR536 pKa = 11.84YY537 pKa = 8.66GGGRR541 pKa = 11.84GRR543 pKa = 11.84RR544 pKa = 11.84WNDD547 pKa = 2.87NQQQQQAQPGRR558 pKa = 11.84SSDD561 pKa = 3.6QPRR564 pKa = 11.84SQSQQPQPEE573 pKa = 3.9ARR575 pKa = 11.84GDD577 pKa = 3.46QSRR580 pKa = 11.84TSGAGRR586 pKa = 11.84GQQGRR591 pKa = 11.84GNQNRR596 pKa = 11.84NQRR599 pKa = 11.84RR600 pKa = 11.84ADD602 pKa = 3.49ANNTRR607 pKa = 11.84NVDD610 pKa = 3.53TVTATTTSSSTASSGQNGSSTTPPASGSRR639 pKa = 11.84NQGDD643 pKa = 3.19
Molecular weight: 69.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3454 |
298 |
1143 |
690.8 |
78.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.906 ± 0.469 | 1.477 ± 0.458 |
4.372 ± 0.339 | 5.472 ± 0.64 |
2.606 ± 0.423 | 5.906 ± 1.318 |
2.403 ± 0.249 | 6.514 ± 0.611 |
5.646 ± 1.468 | 9.641 ± 0.527 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.824 ± 0.417 | 5.067 ± 0.335 |
7.267 ± 0.81 | 6.34 ± 1.227 |
5.269 ± 1.04 | 6.601 ± 0.386 |
5.646 ± 0.559 | 6.022 ± 0.414 |
2.142 ± 0.241 | 3.88 ± 0.581 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
