
Parabacteroides johnsonii CL02T12C29
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides johnsonii
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
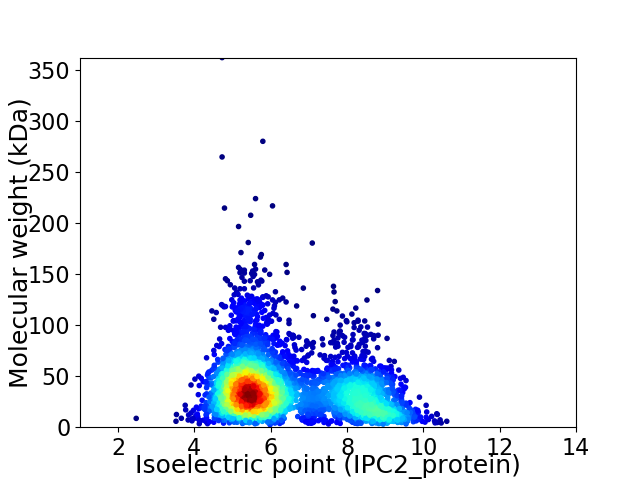
Virtual 2D-PAGE plot for 3736 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
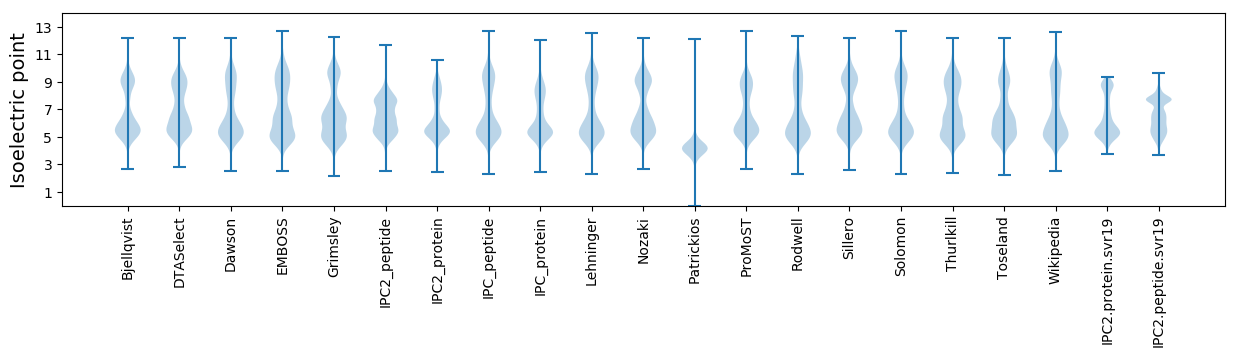
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K5Y9I6|K5Y9I6_9BACT PfkB domain-containing protein OS=Parabacteroides johnsonii CL02T12C29 OX=999419 GN=HMPREF1077_01852 PE=4 SV=1
MM1 pKa = 7.66EE2 pKa = 4.97NLRR5 pKa = 11.84NLSRR9 pKa = 11.84WGMFAFLSVALCTSFVGCSDD29 pKa = 5.24DD30 pKa = 5.71DD31 pKa = 4.06PDD33 pKa = 4.12YY34 pKa = 11.93SNVTPPTVSVSHH46 pKa = 6.57SISGRR51 pKa = 11.84VTGMDD56 pKa = 3.61GNGLSATVSMNGEE69 pKa = 4.23STQTGADD76 pKa = 3.41GTFTFDD82 pKa = 4.93DD83 pKa = 3.97VDD85 pKa = 3.84AGSYY89 pKa = 9.32TLTASAPGKK98 pKa = 8.92QSKK101 pKa = 7.79EE102 pKa = 4.09TTVTVAEE109 pKa = 4.54SGSGANVVWNVSLPNEE125 pKa = 4.02GTTIEE130 pKa = 4.23VAANGDD136 pKa = 3.45TEE138 pKa = 5.54ANVTSEE144 pKa = 4.29TIEE147 pKa = 4.38GNDD150 pKa = 3.25EE151 pKa = 4.09GAVTVAVTVPEE162 pKa = 4.27TAEE165 pKa = 4.12LPAGSSIVVTPLYY178 pKa = 10.43TLDD181 pKa = 3.42EE182 pKa = 4.69AEE184 pKa = 4.44ANTRR188 pKa = 11.84ATSRR192 pKa = 11.84AAEE195 pKa = 4.08SVMLIGTSVACTDD208 pKa = 3.59ANATLSSPIEE218 pKa = 3.72LAYY221 pKa = 10.67DD222 pKa = 3.59VDD224 pKa = 4.43AEE226 pKa = 4.44VAQSITAQKK235 pKa = 9.47YY236 pKa = 8.92VNGQWVDD243 pKa = 3.03AEE245 pKa = 4.43YY246 pKa = 9.75TVEE249 pKa = 4.23GGQVIVFADD258 pKa = 3.37QFTSYY263 pKa = 10.01TLLFGADD270 pKa = 3.62VTSSSTSTPLAFEE283 pKa = 3.63QDD285 pKa = 3.31LWDD288 pKa = 3.83NLYY291 pKa = 11.1GSGDD295 pKa = 3.61MTVGSASFTYY305 pKa = 10.36HH306 pKa = 6.63IGTEE310 pKa = 4.13ITSSGTSRR318 pKa = 11.84ITAYY322 pKa = 10.64LIEE325 pKa = 5.08ILARR329 pKa = 11.84IAGAGVTTATGTYY342 pKa = 8.51PINVTLPVGTALHH355 pKa = 5.82VAGTQQVTTLTVSALNRR372 pKa = 11.84SVSGRR377 pKa = 11.84QYY379 pKa = 11.17GDD381 pKa = 3.19VAVVTTSYY389 pKa = 11.08NRR391 pKa = 11.84NHH393 pKa = 6.77NGGTSDD399 pKa = 3.22NN400 pKa = 3.97
MM1 pKa = 7.66EE2 pKa = 4.97NLRR5 pKa = 11.84NLSRR9 pKa = 11.84WGMFAFLSVALCTSFVGCSDD29 pKa = 5.24DD30 pKa = 5.71DD31 pKa = 4.06PDD33 pKa = 4.12YY34 pKa = 11.93SNVTPPTVSVSHH46 pKa = 6.57SISGRR51 pKa = 11.84VTGMDD56 pKa = 3.61GNGLSATVSMNGEE69 pKa = 4.23STQTGADD76 pKa = 3.41GTFTFDD82 pKa = 4.93DD83 pKa = 3.97VDD85 pKa = 3.84AGSYY89 pKa = 9.32TLTASAPGKK98 pKa = 8.92QSKK101 pKa = 7.79EE102 pKa = 4.09TTVTVAEE109 pKa = 4.54SGSGANVVWNVSLPNEE125 pKa = 4.02GTTIEE130 pKa = 4.23VAANGDD136 pKa = 3.45TEE138 pKa = 5.54ANVTSEE144 pKa = 4.29TIEE147 pKa = 4.38GNDD150 pKa = 3.25EE151 pKa = 4.09GAVTVAVTVPEE162 pKa = 4.27TAEE165 pKa = 4.12LPAGSSIVVTPLYY178 pKa = 10.43TLDD181 pKa = 3.42EE182 pKa = 4.69AEE184 pKa = 4.44ANTRR188 pKa = 11.84ATSRR192 pKa = 11.84AAEE195 pKa = 4.08SVMLIGTSVACTDD208 pKa = 3.59ANATLSSPIEE218 pKa = 3.72LAYY221 pKa = 10.67DD222 pKa = 3.59VDD224 pKa = 4.43AEE226 pKa = 4.44VAQSITAQKK235 pKa = 9.47YY236 pKa = 8.92VNGQWVDD243 pKa = 3.03AEE245 pKa = 4.43YY246 pKa = 9.75TVEE249 pKa = 4.23GGQVIVFADD258 pKa = 3.37QFTSYY263 pKa = 10.01TLLFGADD270 pKa = 3.62VTSSSTSTPLAFEE283 pKa = 3.63QDD285 pKa = 3.31LWDD288 pKa = 3.83NLYY291 pKa = 11.1GSGDD295 pKa = 3.61MTVGSASFTYY305 pKa = 10.36HH306 pKa = 6.63IGTEE310 pKa = 4.13ITSSGTSRR318 pKa = 11.84ITAYY322 pKa = 10.64LIEE325 pKa = 5.08ILARR329 pKa = 11.84IAGAGVTTATGTYY342 pKa = 8.51PINVTLPVGTALHH355 pKa = 5.82VAGTQQVTTLTVSALNRR372 pKa = 11.84SVSGRR377 pKa = 11.84QYY379 pKa = 11.17GDD381 pKa = 3.19VAVVTTSYY389 pKa = 11.08NRR391 pKa = 11.84NHH393 pKa = 6.77NGGTSDD399 pKa = 3.22NN400 pKa = 3.97
Molecular weight: 41.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K6A1X0|K6A1X0_9BACT RNA polymerase sigma-70 factor expansion family 1 OS=Parabacteroides johnsonii CL02T12C29 OX=999419 GN=HMPREF1077_02163 PE=3 SV=1
MM1 pKa = 7.55GFRR4 pKa = 11.84VNRR7 pKa = 11.84RR8 pKa = 11.84TLLLIAGCVWIIAGINILRR27 pKa = 11.84IGIVTWLNDD36 pKa = 2.91AHH38 pKa = 6.52YY39 pKa = 9.65WLFKK43 pKa = 9.85VGEE46 pKa = 4.45AIVVFLLFFQFVFRR60 pKa = 11.84RR61 pKa = 11.84LFIKK65 pKa = 10.19HH66 pKa = 5.93SEE68 pKa = 4.22RR69 pKa = 11.84ISQKK73 pKa = 10.2GEE75 pKa = 3.89KK76 pKa = 9.4NCPFSFFDD84 pKa = 3.48VKK86 pKa = 10.74GWIVMIFMITFGITIRR102 pKa = 11.84HH103 pKa = 5.37FQLLPNSFISVFYY116 pKa = 10.17TGLSSALIITGILFLIRR133 pKa = 11.84AYY135 pKa = 10.61SHH137 pKa = 6.03TFNRR141 pKa = 11.84SS142 pKa = 2.53
MM1 pKa = 7.55GFRR4 pKa = 11.84VNRR7 pKa = 11.84RR8 pKa = 11.84TLLLIAGCVWIIAGINILRR27 pKa = 11.84IGIVTWLNDD36 pKa = 2.91AHH38 pKa = 6.52YY39 pKa = 9.65WLFKK43 pKa = 9.85VGEE46 pKa = 4.45AIVVFLLFFQFVFRR60 pKa = 11.84RR61 pKa = 11.84LFIKK65 pKa = 10.19HH66 pKa = 5.93SEE68 pKa = 4.22RR69 pKa = 11.84ISQKK73 pKa = 10.2GEE75 pKa = 3.89KK76 pKa = 9.4NCPFSFFDD84 pKa = 3.48VKK86 pKa = 10.74GWIVMIFMITFGITIRR102 pKa = 11.84HH103 pKa = 5.37FQLLPNSFISVFYY116 pKa = 10.17TGLSSALIITGILFLIRR133 pKa = 11.84AYY135 pKa = 10.61SHH137 pKa = 6.03TFNRR141 pKa = 11.84SS142 pKa = 2.53
Molecular weight: 16.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1374269 |
22 |
3256 |
367.8 |
41.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.07 ± 0.041 | 1.186 ± 0.015 |
5.599 ± 0.024 | 6.639 ± 0.035 |
4.646 ± 0.024 | 6.915 ± 0.038 |
1.825 ± 0.017 | 6.827 ± 0.037 |
6.611 ± 0.031 | 9.205 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.707 ± 0.017 | 4.982 ± 0.036 |
3.838 ± 0.024 | 3.395 ± 0.02 |
4.706 ± 0.03 | 6.057 ± 0.03 |
5.56 ± 0.024 | 6.565 ± 0.032 |
1.263 ± 0.018 | 4.404 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
