
Kockovaella imperatae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cuniculitremaceae; Kockovaella
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
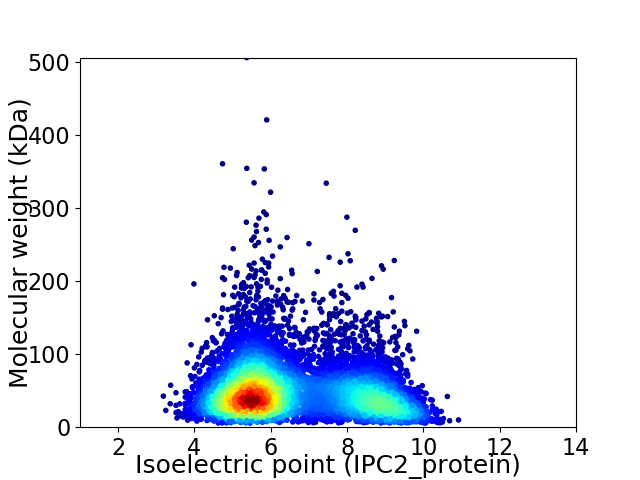
Virtual 2D-PAGE plot for 7388 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y1ULI1|A0A1Y1ULI1_9TREE Uncharacterized protein OS=Kockovaella imperatae OX=4999 GN=BD324DRAFT_617509 PE=4 SV=1
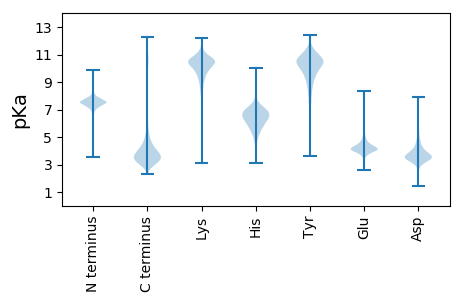
MM1 pKa = 7.28SACSSPDD8 pKa = 3.45LEE10 pKa = 4.45IVTPALTVEE19 pKa = 3.99GDD21 pKa = 4.23FYY23 pKa = 11.61CNEE26 pKa = 4.14DD27 pKa = 3.35DD28 pKa = 4.57VYY30 pKa = 11.24EE31 pKa = 4.25LGYY34 pKa = 10.47EE35 pKa = 4.13YY36 pKa = 10.7EE37 pKa = 4.4SDD39 pKa = 3.77SEE41 pKa = 4.55SEE43 pKa = 4.44SEE45 pKa = 4.45SEE47 pKa = 3.99SHH49 pKa = 6.87AGVGFDD55 pKa = 4.22LFRR58 pKa = 11.84FPSIGFMPSIGQVLDD73 pKa = 4.39SIEE76 pKa = 4.51CPTVEE81 pKa = 4.33KK82 pKa = 8.45TTAVDD87 pKa = 3.08THH89 pKa = 7.9SFDD92 pKa = 6.02DD93 pKa = 4.11VTTHH97 pKa = 7.98DD98 pKa = 5.43DD99 pKa = 4.98DD100 pKa = 6.8DD101 pKa = 6.86DD102 pKa = 5.15NDD104 pKa = 3.75EE105 pKa = 5.57DD106 pKa = 4.89GDD108 pKa = 3.95PSYY111 pKa = 11.4LEE113 pKa = 3.76EE114 pKa = 4.08NARR117 pKa = 11.84DD118 pKa = 4.15FIGPRR123 pKa = 11.84TLAFQAVDD131 pKa = 3.67EE132 pKa = 4.53SWEE135 pKa = 4.18HH136 pKa = 6.84FIGPHH141 pKa = 4.82SKK143 pKa = 9.69MYY145 pKa = 10.76LEE147 pKa = 4.7WIAYY151 pKa = 9.37NEE153 pKa = 4.1AVDD156 pKa = 3.86AAEE159 pKa = 4.14KK160 pKa = 10.19EE161 pKa = 4.59YY162 pKa = 11.29VFTRR166 pKa = 11.84VMEE169 pKa = 4.52GSEE172 pKa = 3.68WTYY175 pKa = 11.6NEE177 pKa = 4.05SDD179 pKa = 3.98ADD181 pKa = 4.22FIGPKK186 pKa = 7.3TQEE189 pKa = 3.46HH190 pKa = 6.46HH191 pKa = 7.02ALDD194 pKa = 3.95EE195 pKa = 4.58ASTGFIGPVSFGGTLLKK212 pKa = 10.74LLEE215 pKa = 4.39PFIEE219 pKa = 4.49AALAADD225 pKa = 3.76EE226 pKa = 4.61EE227 pKa = 4.74EE228 pKa = 4.53EE229 pKa = 4.78KK230 pKa = 11.18EE231 pKa = 3.99GDD233 pKa = 3.25TSFRR237 pKa = 11.84QEE239 pKa = 3.34GAKK242 pKa = 10.54YY243 pKa = 10.64VDD245 pKa = 4.69AEE247 pKa = 4.29DD248 pKa = 3.68QAEE251 pKa = 4.22MEE253 pKa = 4.33HH254 pKa = 7.17VEE256 pKa = 4.61LIGAEE261 pKa = 3.94SDD263 pKa = 4.08LEE265 pKa = 4.49GEE267 pKa = 4.27GSGWVGCVIAA277 pKa = 5.6
MM1 pKa = 7.28SACSSPDD8 pKa = 3.45LEE10 pKa = 4.45IVTPALTVEE19 pKa = 3.99GDD21 pKa = 4.23FYY23 pKa = 11.61CNEE26 pKa = 4.14DD27 pKa = 3.35DD28 pKa = 4.57VYY30 pKa = 11.24EE31 pKa = 4.25LGYY34 pKa = 10.47EE35 pKa = 4.13YY36 pKa = 10.7EE37 pKa = 4.4SDD39 pKa = 3.77SEE41 pKa = 4.55SEE43 pKa = 4.44SEE45 pKa = 4.45SEE47 pKa = 3.99SHH49 pKa = 6.87AGVGFDD55 pKa = 4.22LFRR58 pKa = 11.84FPSIGFMPSIGQVLDD73 pKa = 4.39SIEE76 pKa = 4.51CPTVEE81 pKa = 4.33KK82 pKa = 8.45TTAVDD87 pKa = 3.08THH89 pKa = 7.9SFDD92 pKa = 6.02DD93 pKa = 4.11VTTHH97 pKa = 7.98DD98 pKa = 5.43DD99 pKa = 4.98DD100 pKa = 6.8DD101 pKa = 6.86DD102 pKa = 5.15NDD104 pKa = 3.75EE105 pKa = 5.57DD106 pKa = 4.89GDD108 pKa = 3.95PSYY111 pKa = 11.4LEE113 pKa = 3.76EE114 pKa = 4.08NARR117 pKa = 11.84DD118 pKa = 4.15FIGPRR123 pKa = 11.84TLAFQAVDD131 pKa = 3.67EE132 pKa = 4.53SWEE135 pKa = 4.18HH136 pKa = 6.84FIGPHH141 pKa = 4.82SKK143 pKa = 9.69MYY145 pKa = 10.76LEE147 pKa = 4.7WIAYY151 pKa = 9.37NEE153 pKa = 4.1AVDD156 pKa = 3.86AAEE159 pKa = 4.14KK160 pKa = 10.19EE161 pKa = 4.59YY162 pKa = 11.29VFTRR166 pKa = 11.84VMEE169 pKa = 4.52GSEE172 pKa = 3.68WTYY175 pKa = 11.6NEE177 pKa = 4.05SDD179 pKa = 3.98ADD181 pKa = 4.22FIGPKK186 pKa = 7.3TQEE189 pKa = 3.46HH190 pKa = 6.46HH191 pKa = 7.02ALDD194 pKa = 3.95EE195 pKa = 4.58ASTGFIGPVSFGGTLLKK212 pKa = 10.74LLEE215 pKa = 4.39PFIEE219 pKa = 4.49AALAADD225 pKa = 3.76EE226 pKa = 4.61EE227 pKa = 4.74EE228 pKa = 4.53EE229 pKa = 4.78KK230 pKa = 11.18EE231 pKa = 3.99GDD233 pKa = 3.25TSFRR237 pKa = 11.84QEE239 pKa = 3.34GAKK242 pKa = 10.54YY243 pKa = 10.64VDD245 pKa = 4.69AEE247 pKa = 4.29DD248 pKa = 3.68QAEE251 pKa = 4.22MEE253 pKa = 4.33HH254 pKa = 7.17VEE256 pKa = 4.61LIGAEE261 pKa = 3.94SDD263 pKa = 4.08LEE265 pKa = 4.49GEE267 pKa = 4.27GSGWVGCVIAA277 pKa = 5.6
Molecular weight: 30.62 kDa
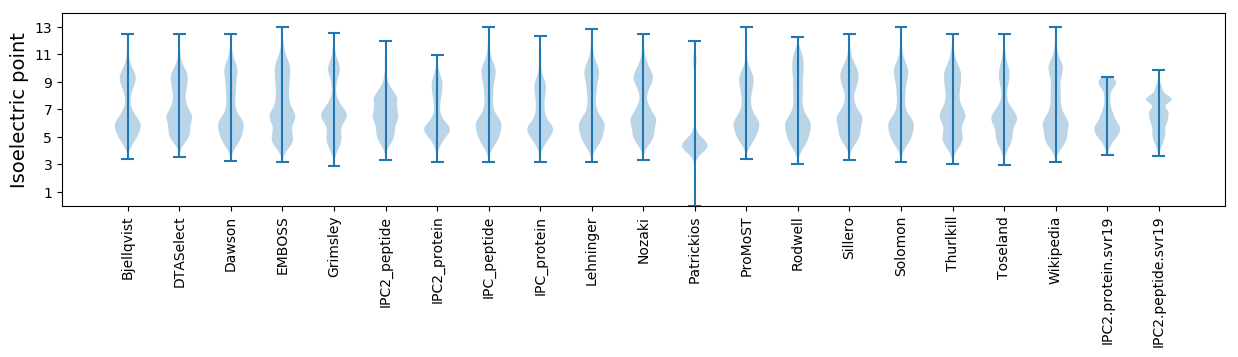
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y1UG68|A0A1Y1UG68_9TREE Uncharacterized protein OS=Kockovaella imperatae OX=4999 GN=BD324DRAFT_629123 PE=4 SV=1
SS1 pKa = 6.16TLRR4 pKa = 11.84ASLIPRR10 pKa = 11.84ITSLLRR16 pKa = 11.84PTLSSPIPSALLPHH30 pKa = 6.85NSPLDD35 pKa = 3.84LGQVRR40 pKa = 11.84FGSKK44 pKa = 10.05GGNTYY49 pKa = 10.38QPSQRR54 pKa = 11.84KK55 pKa = 8.78RR56 pKa = 11.84KK57 pKa = 8.94RR58 pKa = 11.84KK59 pKa = 9.42HH60 pKa = 5.72GFLSRR65 pKa = 11.84IKK67 pKa = 8.35TAKK70 pKa = 9.0GRR72 pKa = 11.84KK73 pKa = 7.91ILTRR77 pKa = 11.84RR78 pKa = 11.84MVKK81 pKa = 9.45GRR83 pKa = 11.84KK84 pKa = 8.69FLSHH88 pKa = 6.98
SS1 pKa = 6.16TLRR4 pKa = 11.84ASLIPRR10 pKa = 11.84ITSLLRR16 pKa = 11.84PTLSSPIPSALLPHH30 pKa = 6.85NSPLDD35 pKa = 3.84LGQVRR40 pKa = 11.84FGSKK44 pKa = 10.05GGNTYY49 pKa = 10.38QPSQRR54 pKa = 11.84KK55 pKa = 8.78RR56 pKa = 11.84KK57 pKa = 8.94RR58 pKa = 11.84KK59 pKa = 9.42HH60 pKa = 5.72GFLSRR65 pKa = 11.84IKK67 pKa = 8.35TAKK70 pKa = 9.0GRR72 pKa = 11.84KK73 pKa = 7.91ILTRR77 pKa = 11.84RR78 pKa = 11.84MVKK81 pKa = 9.45GRR83 pKa = 11.84KK84 pKa = 8.69FLSHH88 pKa = 6.98
Molecular weight: 9.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3644630 |
50 |
4468 |
493.3 |
54.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.622 ± 0.023 | 1.066 ± 0.009 |
5.722 ± 0.021 | 6.035 ± 0.027 |
3.371 ± 0.018 | 7.172 ± 0.027 |
2.406 ± 0.012 | 4.751 ± 0.021 |
4.604 ± 0.026 | 8.991 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.235 ± 0.01 | 3.182 ± 0.015 |
6.572 ± 0.034 | 3.828 ± 0.017 |
6.311 ± 0.026 | 9.261 ± 0.041 |
5.918 ± 0.016 | 6.034 ± 0.018 |
1.444 ± 0.01 | 2.476 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
