
Wenling tombus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.89
Get precalculated fractions of proteins
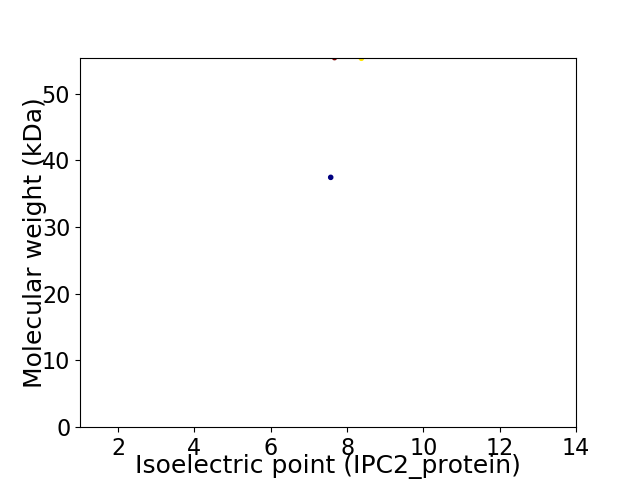
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGU4|A0A1L3KGU4_9VIRU CCHC-type domain-containing protein OS=Wenling tombus-like virus 1 OX=1923543 PE=4 SV=1
MM1 pKa = 6.54TQPRR5 pKa = 11.84PSHH8 pKa = 5.97TNNSNKK14 pKa = 8.28TMRR17 pKa = 11.84CYY19 pKa = 10.17RR20 pKa = 11.84CNEE23 pKa = 3.98EE24 pKa = 3.51GHH26 pKa = 6.43IARR29 pKa = 11.84DD30 pKa = 3.64CVPQRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GGRR41 pKa = 11.84GSGQAQRR48 pKa = 11.84AVRR51 pKa = 11.84ASDD54 pKa = 3.03EE55 pKa = 4.35RR56 pKa = 11.84SRR58 pKa = 11.84HH59 pKa = 3.45VHH61 pKa = 6.02SCVKK65 pKa = 10.49CCGLYY70 pKa = 10.41SHH72 pKa = 7.11IHH74 pKa = 6.43PYY76 pKa = 9.25NHH78 pKa = 7.16EE79 pKa = 4.05DD80 pKa = 3.41HH81 pKa = 6.55EE82 pKa = 4.3QRR84 pKa = 11.84PYY86 pKa = 9.28QCPYY90 pKa = 8.01PTCEE94 pKa = 3.96WYY96 pKa = 9.96IGSDD100 pKa = 2.94TAEE103 pKa = 4.26EE104 pKa = 4.62YY105 pKa = 8.17RR106 pKa = 11.84TKK108 pKa = 10.69PQMLEE113 pKa = 4.34PIPTPTPTAPPTPEE127 pKa = 4.1PVDD130 pKa = 3.75PLVGPPIDD138 pKa = 4.64EE139 pKa = 4.52EE140 pKa = 4.32PADD143 pKa = 3.81EE144 pKa = 4.22STPKK148 pKa = 10.67GFSSFNDD155 pKa = 3.66LKK157 pKa = 11.23DD158 pKa = 3.25HH159 pKa = 6.97LYY161 pKa = 10.7AYY163 pKa = 7.69LTKK166 pKa = 10.56QSAWTEE172 pKa = 3.73RR173 pKa = 11.84TVLNRR178 pKa = 11.84RR179 pKa = 11.84SMVQRR184 pKa = 11.84AEE186 pKa = 3.62TWLVSKK192 pKa = 10.62KK193 pKa = 10.62VEE195 pKa = 4.22SEE197 pKa = 4.09VLRR200 pKa = 11.84TQVIQAVIDD209 pKa = 3.79TVDD212 pKa = 2.81IMNPLEE218 pKa = 4.17VAVTSAAQNVEE229 pKa = 3.24WRR231 pKa = 11.84NQHH234 pKa = 5.18IAANRR239 pKa = 11.84AAAGWLYY246 pKa = 10.35QPPAVGWSLFTKK258 pKa = 10.17FLIITLLFMTSIIAVCLIGPGVVANSIASLAGHH291 pKa = 7.0AVAKK295 pKa = 8.87TVSINISFLLANAAIMAWRR314 pKa = 11.84GRR316 pKa = 11.84EE317 pKa = 3.89RR318 pKa = 11.84APHH321 pKa = 5.11EE322 pKa = 4.47TGNSAVFTSGNMGRR336 pKa = 4.34
MM1 pKa = 6.54TQPRR5 pKa = 11.84PSHH8 pKa = 5.97TNNSNKK14 pKa = 8.28TMRR17 pKa = 11.84CYY19 pKa = 10.17RR20 pKa = 11.84CNEE23 pKa = 3.98EE24 pKa = 3.51GHH26 pKa = 6.43IARR29 pKa = 11.84DD30 pKa = 3.64CVPQRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GGRR41 pKa = 11.84GSGQAQRR48 pKa = 11.84AVRR51 pKa = 11.84ASDD54 pKa = 3.03EE55 pKa = 4.35RR56 pKa = 11.84SRR58 pKa = 11.84HH59 pKa = 3.45VHH61 pKa = 6.02SCVKK65 pKa = 10.49CCGLYY70 pKa = 10.41SHH72 pKa = 7.11IHH74 pKa = 6.43PYY76 pKa = 9.25NHH78 pKa = 7.16EE79 pKa = 4.05DD80 pKa = 3.41HH81 pKa = 6.55EE82 pKa = 4.3QRR84 pKa = 11.84PYY86 pKa = 9.28QCPYY90 pKa = 8.01PTCEE94 pKa = 3.96WYY96 pKa = 9.96IGSDD100 pKa = 2.94TAEE103 pKa = 4.26EE104 pKa = 4.62YY105 pKa = 8.17RR106 pKa = 11.84TKK108 pKa = 10.69PQMLEE113 pKa = 4.34PIPTPTPTAPPTPEE127 pKa = 4.1PVDD130 pKa = 3.75PLVGPPIDD138 pKa = 4.64EE139 pKa = 4.52EE140 pKa = 4.32PADD143 pKa = 3.81EE144 pKa = 4.22STPKK148 pKa = 10.67GFSSFNDD155 pKa = 3.66LKK157 pKa = 11.23DD158 pKa = 3.25HH159 pKa = 6.97LYY161 pKa = 10.7AYY163 pKa = 7.69LTKK166 pKa = 10.56QSAWTEE172 pKa = 3.73RR173 pKa = 11.84TVLNRR178 pKa = 11.84RR179 pKa = 11.84SMVQRR184 pKa = 11.84AEE186 pKa = 3.62TWLVSKK192 pKa = 10.62KK193 pKa = 10.62VEE195 pKa = 4.22SEE197 pKa = 4.09VLRR200 pKa = 11.84TQVIQAVIDD209 pKa = 3.79TVDD212 pKa = 2.81IMNPLEE218 pKa = 4.17VAVTSAAQNVEE229 pKa = 3.24WRR231 pKa = 11.84NQHH234 pKa = 5.18IAANRR239 pKa = 11.84AAAGWLYY246 pKa = 10.35QPPAVGWSLFTKK258 pKa = 10.17FLIITLLFMTSIIAVCLIGPGVVANSIASLAGHH291 pKa = 7.0AVAKK295 pKa = 8.87TVSINISFLLANAAIMAWRR314 pKa = 11.84GRR316 pKa = 11.84EE317 pKa = 3.89RR318 pKa = 11.84APHH321 pKa = 5.11EE322 pKa = 4.47TGNSAVFTSGNMGRR336 pKa = 4.34
Molecular weight: 37.46 kDa
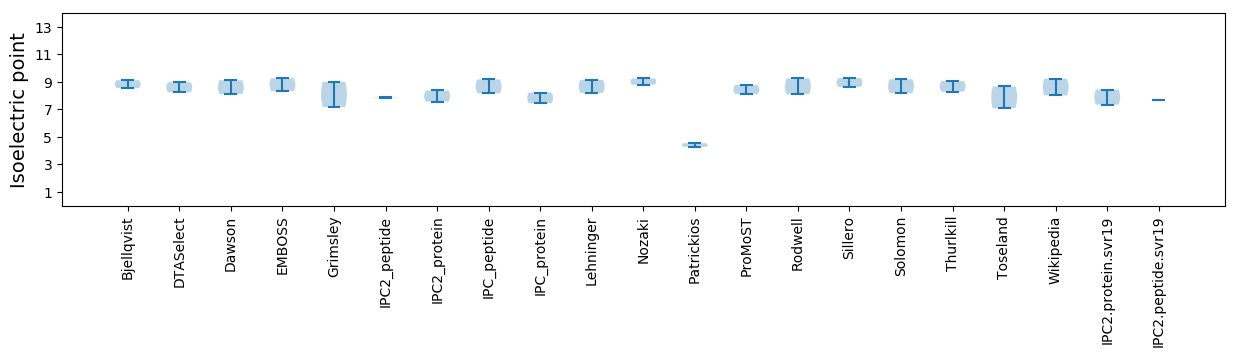
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGU4|A0A1L3KGU4_9VIRU CCHC-type domain-containing protein OS=Wenling tombus-like virus 1 OX=1923543 PE=4 SV=1
MM1 pKa = 7.46WNSIPGCYY9 pKa = 9.98NPFVHH14 pKa = 6.66SNCVRR19 pKa = 11.84NEE21 pKa = 3.76YY22 pKa = 10.61VAMRR26 pKa = 11.84DD27 pKa = 3.19RR28 pKa = 11.84VVGKK32 pKa = 10.51VPVPTEE38 pKa = 3.53RR39 pKa = 11.84GVRR42 pKa = 11.84LLRR45 pKa = 11.84RR46 pKa = 11.84YY47 pKa = 9.18ARR49 pKa = 11.84KK50 pKa = 9.12LVKK53 pKa = 10.34RR54 pKa = 11.84LGKK57 pKa = 8.8TDD59 pKa = 4.15EE60 pKa = 3.9ISYY63 pKa = 10.29DD64 pKa = 3.57QVIDD68 pKa = 3.95HH69 pKa = 5.07YY70 pKa = 10.85HH71 pKa = 4.76GRR73 pKa = 11.84KK74 pKa = 8.26RR75 pKa = 11.84KK76 pKa = 9.36RR77 pKa = 11.84YY78 pKa = 7.77EE79 pKa = 3.59AAKK82 pKa = 10.24EE83 pKa = 4.08SLGRR87 pKa = 11.84LALTKK92 pKa = 10.33NDD94 pKa = 4.24SIIKK98 pKa = 10.49AFVKK102 pKa = 10.6CEE104 pKa = 3.85KK105 pKa = 10.3FNPADD110 pKa = 3.67KK111 pKa = 10.98VNPAPRR117 pKa = 11.84MIQARR122 pKa = 11.84NARR125 pKa = 11.84YY126 pKa = 9.28NLSIARR132 pKa = 11.84WLRR135 pKa = 11.84PIEE138 pKa = 3.83HH139 pKa = 7.31AIYY142 pKa = 10.24RR143 pKa = 11.84LKK145 pKa = 11.01SKK147 pKa = 9.52LTGLPIVGKK156 pKa = 10.24GRR158 pKa = 11.84SLNEE162 pKa = 3.75RR163 pKa = 11.84AEE165 pKa = 4.1LLAKK169 pKa = 10.02KK170 pKa = 10.23FNYY173 pKa = 9.13FKK175 pKa = 11.3NPVVYY180 pKa = 10.7SLDD183 pKa = 3.29ASRR186 pKa = 11.84WDD188 pKa = 3.29QHH190 pKa = 7.95CDD192 pKa = 2.87IKK194 pKa = 11.15LLEE197 pKa = 4.18VEE199 pKa = 3.97HH200 pKa = 6.57WIYY203 pKa = 11.08RR204 pKa = 11.84AMNGSSEE211 pKa = 4.13FATLLQQQLYY221 pKa = 8.59NKK223 pKa = 9.65CFTEE227 pKa = 4.11HH228 pKa = 6.61GIAYY232 pKa = 6.12KK233 pKa = 10.43TKK235 pKa = 8.12GKK237 pKa = 10.48RR238 pKa = 11.84MSGDD242 pKa = 3.08VNTALGNCLLAILLAYY258 pKa = 9.71VVLVDD263 pKa = 3.95FLKK266 pKa = 11.06LKK268 pKa = 8.86EE269 pKa = 4.45TEE271 pKa = 4.84FEE273 pKa = 4.42LLDD276 pKa = 5.14DD277 pKa = 5.25GDD279 pKa = 4.37DD280 pKa = 3.64LLVIVEE286 pKa = 4.39QKK288 pKa = 10.72DD289 pKa = 3.39EE290 pKa = 4.17HH291 pKa = 7.42RR292 pKa = 11.84LEE294 pKa = 4.9GIKK297 pKa = 9.75EE298 pKa = 3.95AYY300 pKa = 10.38LEE302 pKa = 4.23VGHH305 pKa = 6.54EE306 pKa = 3.94IKK308 pKa = 10.85LEE310 pKa = 3.8NRR312 pKa = 11.84ATVLEE317 pKa = 4.14DD318 pKa = 4.52VEE320 pKa = 4.78WCQHH324 pKa = 5.75RR325 pKa = 11.84PVHH328 pKa = 5.88TPNGWRR334 pKa = 11.84FVPNWKK340 pKa = 9.77KK341 pKa = 10.52VLSSTTCDD349 pKa = 3.19SKK351 pKa = 11.35HH352 pKa = 5.41WMHH355 pKa = 6.68EE356 pKa = 3.76NLRR359 pKa = 11.84PSLGHH364 pKa = 6.27TMGKK368 pKa = 9.61CLLSMYY374 pKa = 9.92TGIPVLQEE382 pKa = 3.54YY383 pKa = 10.37CQFLIQQGNKK393 pKa = 9.19DD394 pKa = 3.82AKK396 pKa = 10.18CLKK399 pKa = 10.16DD400 pKa = 3.5HH401 pKa = 6.5YY402 pKa = 11.4LMDD405 pKa = 4.11RR406 pKa = 11.84AYY408 pKa = 9.06TNGGATTDD416 pKa = 3.64TTPVDD421 pKa = 3.45PEE423 pKa = 4.2TRR425 pKa = 11.84LSFQRR430 pKa = 11.84AFGMTVQEE438 pKa = 4.1QLDD441 pKa = 3.65IEE443 pKa = 4.4TRR445 pKa = 11.84IRR447 pKa = 11.84GLTLPTTTLRR457 pKa = 11.84ITASEE462 pKa = 4.19VLPGWKK468 pKa = 8.67WEE470 pKa = 4.12YY471 pKa = 11.18YY472 pKa = 9.57PGTEE476 pKa = 4.58AGLGG480 pKa = 3.55
MM1 pKa = 7.46WNSIPGCYY9 pKa = 9.98NPFVHH14 pKa = 6.66SNCVRR19 pKa = 11.84NEE21 pKa = 3.76YY22 pKa = 10.61VAMRR26 pKa = 11.84DD27 pKa = 3.19RR28 pKa = 11.84VVGKK32 pKa = 10.51VPVPTEE38 pKa = 3.53RR39 pKa = 11.84GVRR42 pKa = 11.84LLRR45 pKa = 11.84RR46 pKa = 11.84YY47 pKa = 9.18ARR49 pKa = 11.84KK50 pKa = 9.12LVKK53 pKa = 10.34RR54 pKa = 11.84LGKK57 pKa = 8.8TDD59 pKa = 4.15EE60 pKa = 3.9ISYY63 pKa = 10.29DD64 pKa = 3.57QVIDD68 pKa = 3.95HH69 pKa = 5.07YY70 pKa = 10.85HH71 pKa = 4.76GRR73 pKa = 11.84KK74 pKa = 8.26RR75 pKa = 11.84KK76 pKa = 9.36RR77 pKa = 11.84YY78 pKa = 7.77EE79 pKa = 3.59AAKK82 pKa = 10.24EE83 pKa = 4.08SLGRR87 pKa = 11.84LALTKK92 pKa = 10.33NDD94 pKa = 4.24SIIKK98 pKa = 10.49AFVKK102 pKa = 10.6CEE104 pKa = 3.85KK105 pKa = 10.3FNPADD110 pKa = 3.67KK111 pKa = 10.98VNPAPRR117 pKa = 11.84MIQARR122 pKa = 11.84NARR125 pKa = 11.84YY126 pKa = 9.28NLSIARR132 pKa = 11.84WLRR135 pKa = 11.84PIEE138 pKa = 3.83HH139 pKa = 7.31AIYY142 pKa = 10.24RR143 pKa = 11.84LKK145 pKa = 11.01SKK147 pKa = 9.52LTGLPIVGKK156 pKa = 10.24GRR158 pKa = 11.84SLNEE162 pKa = 3.75RR163 pKa = 11.84AEE165 pKa = 4.1LLAKK169 pKa = 10.02KK170 pKa = 10.23FNYY173 pKa = 9.13FKK175 pKa = 11.3NPVVYY180 pKa = 10.7SLDD183 pKa = 3.29ASRR186 pKa = 11.84WDD188 pKa = 3.29QHH190 pKa = 7.95CDD192 pKa = 2.87IKK194 pKa = 11.15LLEE197 pKa = 4.18VEE199 pKa = 3.97HH200 pKa = 6.57WIYY203 pKa = 11.08RR204 pKa = 11.84AMNGSSEE211 pKa = 4.13FATLLQQQLYY221 pKa = 8.59NKK223 pKa = 9.65CFTEE227 pKa = 4.11HH228 pKa = 6.61GIAYY232 pKa = 6.12KK233 pKa = 10.43TKK235 pKa = 8.12GKK237 pKa = 10.48RR238 pKa = 11.84MSGDD242 pKa = 3.08VNTALGNCLLAILLAYY258 pKa = 9.71VVLVDD263 pKa = 3.95FLKK266 pKa = 11.06LKK268 pKa = 8.86EE269 pKa = 4.45TEE271 pKa = 4.84FEE273 pKa = 4.42LLDD276 pKa = 5.14DD277 pKa = 5.25GDD279 pKa = 4.37DD280 pKa = 3.64LLVIVEE286 pKa = 4.39QKK288 pKa = 10.72DD289 pKa = 3.39EE290 pKa = 4.17HH291 pKa = 7.42RR292 pKa = 11.84LEE294 pKa = 4.9GIKK297 pKa = 9.75EE298 pKa = 3.95AYY300 pKa = 10.38LEE302 pKa = 4.23VGHH305 pKa = 6.54EE306 pKa = 3.94IKK308 pKa = 10.85LEE310 pKa = 3.8NRR312 pKa = 11.84ATVLEE317 pKa = 4.14DD318 pKa = 4.52VEE320 pKa = 4.78WCQHH324 pKa = 5.75RR325 pKa = 11.84PVHH328 pKa = 5.88TPNGWRR334 pKa = 11.84FVPNWKK340 pKa = 9.77KK341 pKa = 10.52VLSSTTCDD349 pKa = 3.19SKK351 pKa = 11.35HH352 pKa = 5.41WMHH355 pKa = 6.68EE356 pKa = 3.76NLRR359 pKa = 11.84PSLGHH364 pKa = 6.27TMGKK368 pKa = 9.61CLLSMYY374 pKa = 9.92TGIPVLQEE382 pKa = 3.54YY383 pKa = 10.37CQFLIQQGNKK393 pKa = 9.19DD394 pKa = 3.82AKK396 pKa = 10.18CLKK399 pKa = 10.16DD400 pKa = 3.5HH401 pKa = 6.5YY402 pKa = 11.4LMDD405 pKa = 4.11RR406 pKa = 11.84AYY408 pKa = 9.06TNGGATTDD416 pKa = 3.64TTPVDD421 pKa = 3.45PEE423 pKa = 4.2TRR425 pKa = 11.84LSFQRR430 pKa = 11.84AFGMTVQEE438 pKa = 4.1QLDD441 pKa = 3.65IEE443 pKa = 4.4TRR445 pKa = 11.84IRR447 pKa = 11.84GLTLPTTTLRR457 pKa = 11.84ITASEE462 pKa = 4.19VLPGWKK468 pKa = 8.67WEE470 pKa = 4.12YY471 pKa = 11.18YY472 pKa = 9.57PGTEE476 pKa = 4.58AGLGG480 pKa = 3.55
Molecular weight: 55.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
816 |
336 |
480 |
408.0 |
46.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.475 ± 1.214 | 2.451 ± 0.135 |
4.289 ± 0.602 | 6.618 ± 0.218 |
2.451 ± 0.218 | 5.882 ± 0.311 |
3.309 ± 0.156 | 4.902 ± 0.27 |
5.637 ± 1.577 | 8.824 ± 1.878 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.206 ± 0.104 | 4.657 ± 0.062 |
5.637 ± 1.245 | 3.554 ± 0.363 |
7.475 ± 0.156 | 5.392 ± 1.038 |
6.495 ± 0.384 | 6.863 ± 0.166 |
2.083 ± 0.0 | 3.799 ± 0.488 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
