
Beihai shrimp virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.83
Get precalculated fractions of proteins
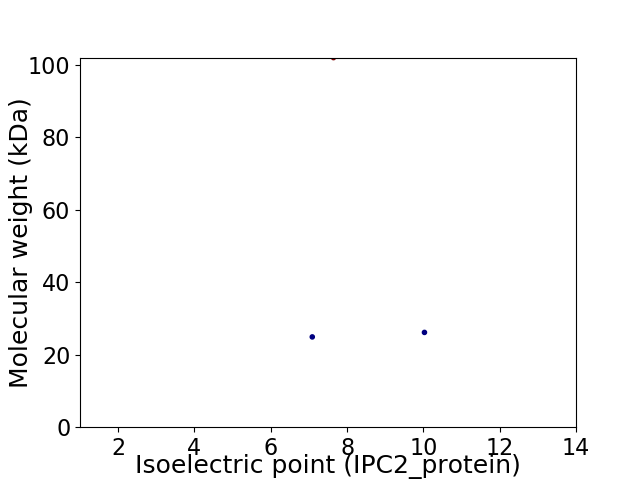
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFH3|A0A1L3KFH3_9VIRU RNA replicase OS=Beihai shrimp virus 6 OX=1922672 PE=3 SV=1
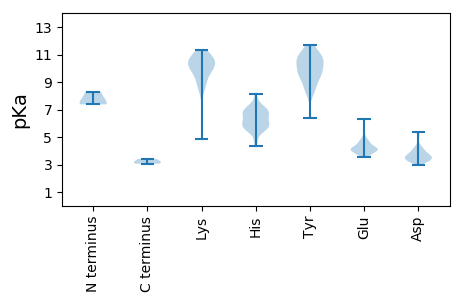
MM1 pKa = 7.38EE2 pKa = 4.61VGVAQRR8 pKa = 11.84TLPSAEE14 pKa = 4.04AEE16 pKa = 4.07HH17 pKa = 8.16VIMKK21 pKa = 10.32RR22 pKa = 11.84HH23 pKa = 5.87DD24 pKa = 3.55PDD26 pKa = 3.35RR27 pKa = 11.84RR28 pKa = 11.84EE29 pKa = 3.67MLINEE34 pKa = 3.93FRR36 pKa = 11.84FGHH39 pKa = 6.84GYY41 pKa = 9.67PGEE44 pKa = 4.31SQFAARR50 pKa = 11.84LFTQSCSFLLEE61 pKa = 4.13LATDD65 pKa = 4.21FTPPEE70 pKa = 3.93GWISDD75 pKa = 3.87LVFEE79 pKa = 4.74FLPHH83 pKa = 6.3CFEE86 pKa = 4.26IVKK89 pKa = 10.3QSIHH93 pKa = 5.69FVEE96 pKa = 5.0VMAPRR101 pKa = 11.84VFWITMTTVARR112 pKa = 11.84ATVEE116 pKa = 4.05TFPEE120 pKa = 4.02YY121 pKa = 10.76LRR123 pKa = 11.84TVLRR127 pKa = 11.84ISIPVAGALQVYY139 pKa = 9.4VDD141 pKa = 3.67KK142 pKa = 11.29QNRR145 pKa = 11.84ALLKK149 pKa = 9.68PGEE152 pKa = 4.07MLFWANISCLTFRR165 pKa = 11.84VLPFQFDD172 pKa = 4.01PLVTDD177 pKa = 4.13PDD179 pKa = 3.69AVVTHH184 pKa = 6.73RR185 pKa = 11.84SISDD189 pKa = 3.34IVRR192 pKa = 11.84HH193 pKa = 5.68HH194 pKa = 6.85PNVRR198 pKa = 11.84HH199 pKa = 5.56HH200 pKa = 5.79RR201 pKa = 11.84HH202 pKa = 6.06RR203 pKa = 11.84YY204 pKa = 8.69GNTIRR209 pKa = 11.84NRR211 pKa = 11.84KK212 pKa = 4.88THH214 pKa = 5.91PP215 pKa = 3.41
MM1 pKa = 7.38EE2 pKa = 4.61VGVAQRR8 pKa = 11.84TLPSAEE14 pKa = 4.04AEE16 pKa = 4.07HH17 pKa = 8.16VIMKK21 pKa = 10.32RR22 pKa = 11.84HH23 pKa = 5.87DD24 pKa = 3.55PDD26 pKa = 3.35RR27 pKa = 11.84RR28 pKa = 11.84EE29 pKa = 3.67MLINEE34 pKa = 3.93FRR36 pKa = 11.84FGHH39 pKa = 6.84GYY41 pKa = 9.67PGEE44 pKa = 4.31SQFAARR50 pKa = 11.84LFTQSCSFLLEE61 pKa = 4.13LATDD65 pKa = 4.21FTPPEE70 pKa = 3.93GWISDD75 pKa = 3.87LVFEE79 pKa = 4.74FLPHH83 pKa = 6.3CFEE86 pKa = 4.26IVKK89 pKa = 10.3QSIHH93 pKa = 5.69FVEE96 pKa = 5.0VMAPRR101 pKa = 11.84VFWITMTTVARR112 pKa = 11.84ATVEE116 pKa = 4.05TFPEE120 pKa = 4.02YY121 pKa = 10.76LRR123 pKa = 11.84TVLRR127 pKa = 11.84ISIPVAGALQVYY139 pKa = 9.4VDD141 pKa = 3.67KK142 pKa = 11.29QNRR145 pKa = 11.84ALLKK149 pKa = 9.68PGEE152 pKa = 4.07MLFWANISCLTFRR165 pKa = 11.84VLPFQFDD172 pKa = 4.01PLVTDD177 pKa = 4.13PDD179 pKa = 3.69AVVTHH184 pKa = 6.73RR185 pKa = 11.84SISDD189 pKa = 3.34IVRR192 pKa = 11.84HH193 pKa = 5.68HH194 pKa = 6.85PNVRR198 pKa = 11.84HH199 pKa = 5.56HH200 pKa = 5.79RR201 pKa = 11.84HH202 pKa = 6.06RR203 pKa = 11.84YY204 pKa = 8.69GNTIRR209 pKa = 11.84NRR211 pKa = 11.84KK212 pKa = 4.88THH214 pKa = 5.91PP215 pKa = 3.41
Molecular weight: 24.92 kDa
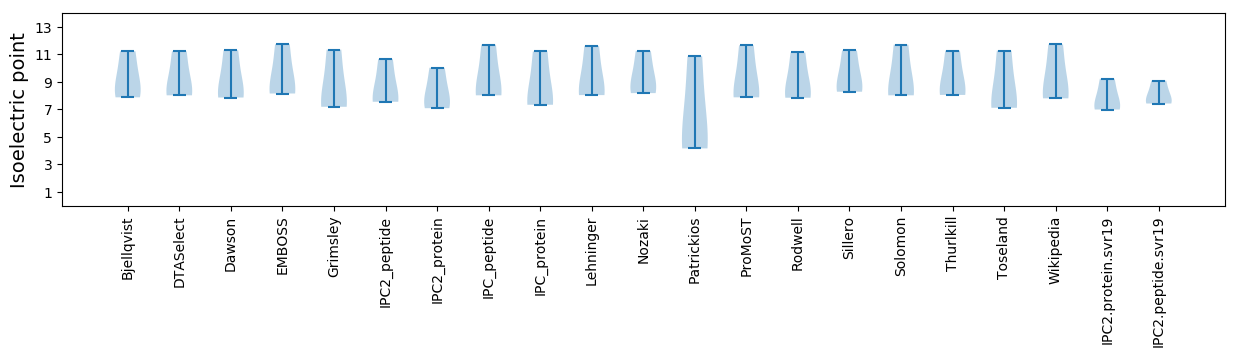
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFJ9|A0A1L3KFJ9_9VIRU Capsid protein OS=Beihai shrimp virus 6 OX=1922672 PE=3 SV=1
MM1 pKa = 7.5TKK3 pKa = 10.2RR4 pKa = 11.84ANRR7 pKa = 11.84RR8 pKa = 11.84LNNAIASLQALTMATRR24 pKa = 11.84TNIRR28 pKa = 11.84AARR31 pKa = 11.84APVRR35 pKa = 11.84SAPQPRR41 pKa = 11.84PGRR44 pKa = 11.84NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84NNRR52 pKa = 11.84GSVPGGYY59 pKa = 6.4TTGSAGPLRR68 pKa = 11.84TSGTNGGVRR77 pKa = 11.84IRR79 pKa = 11.84HH80 pKa = 5.5EE81 pKa = 4.26EE82 pKa = 3.65MFLNLRR88 pKa = 11.84PRR90 pKa = 11.84VAGEE94 pKa = 4.1NIATQIFLPGQSKK107 pKa = 9.0MPVLDD112 pKa = 3.41SFARR116 pKa = 11.84SFDD119 pKa = 3.2RR120 pKa = 11.84YY121 pKa = 9.83RR122 pKa = 11.84IHH124 pKa = 7.53SLTLRR129 pKa = 11.84YY130 pKa = 8.09KK131 pKa = 9.1TASGTTKK138 pKa = 9.65TGSVILGIDD147 pKa = 3.55GAVDD151 pKa = 3.61HH152 pKa = 6.85VPTTIAQVQGLYY164 pKa = 9.91PKK166 pKa = 9.76WRR168 pKa = 11.84GPVWGEE174 pKa = 3.53GAITANIATLMPQRR188 pKa = 11.84WLTTSTDD195 pKa = 2.99VSMLKK200 pKa = 10.31SVAHH204 pKa = 6.27AAFAVVVGLTSAEE217 pKa = 4.04PDD219 pKa = 3.51VSPGEE224 pKa = 4.0IWCSYY229 pKa = 10.29DD230 pKa = 3.39VEE232 pKa = 4.55FMYY235 pKa = 8.68PTGSGNN241 pKa = 3.18
MM1 pKa = 7.5TKK3 pKa = 10.2RR4 pKa = 11.84ANRR7 pKa = 11.84RR8 pKa = 11.84LNNAIASLQALTMATRR24 pKa = 11.84TNIRR28 pKa = 11.84AARR31 pKa = 11.84APVRR35 pKa = 11.84SAPQPRR41 pKa = 11.84PGRR44 pKa = 11.84NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84NNRR52 pKa = 11.84GSVPGGYY59 pKa = 6.4TTGSAGPLRR68 pKa = 11.84TSGTNGGVRR77 pKa = 11.84IRR79 pKa = 11.84HH80 pKa = 5.5EE81 pKa = 4.26EE82 pKa = 3.65MFLNLRR88 pKa = 11.84PRR90 pKa = 11.84VAGEE94 pKa = 4.1NIATQIFLPGQSKK107 pKa = 9.0MPVLDD112 pKa = 3.41SFARR116 pKa = 11.84SFDD119 pKa = 3.2RR120 pKa = 11.84YY121 pKa = 9.83RR122 pKa = 11.84IHH124 pKa = 7.53SLTLRR129 pKa = 11.84YY130 pKa = 8.09KK131 pKa = 9.1TASGTTKK138 pKa = 9.65TGSVILGIDD147 pKa = 3.55GAVDD151 pKa = 3.61HH152 pKa = 6.85VPTTIAQVQGLYY164 pKa = 9.91PKK166 pKa = 9.76WRR168 pKa = 11.84GPVWGEE174 pKa = 3.53GAITANIATLMPQRR188 pKa = 11.84WLTTSTDD195 pKa = 2.99VSMLKK200 pKa = 10.31SVAHH204 pKa = 6.27AAFAVVVGLTSAEE217 pKa = 4.04PDD219 pKa = 3.51VSPGEE224 pKa = 4.0IWCSYY229 pKa = 10.29DD230 pKa = 3.39VEE232 pKa = 4.55FMYY235 pKa = 8.68PTGSGNN241 pKa = 3.18
Molecular weight: 26.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1372 |
215 |
916 |
457.3 |
51.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.09 ± 0.522 | 1.676 ± 0.428 |
5.248 ± 0.917 | 5.102 ± 0.571 |
3.134 ± 1.276 | 7.289 ± 0.973 |
2.843 ± 0.75 | 4.883 ± 0.237 |
4.155 ± 0.936 | 7.07 ± 0.227 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.353 | 3.863 ± 0.331 |
6.122 ± 0.374 | 2.697 ± 0.216 |
8.163 ± 0.789 | 6.997 ± 0.682 |
6.633 ± 0.927 | 8.236 ± 0.34 |
1.603 ± 0.059 | 4.009 ± 0.992 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
