
Anguilla anguilla circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 8.86
Get precalculated fractions of proteins
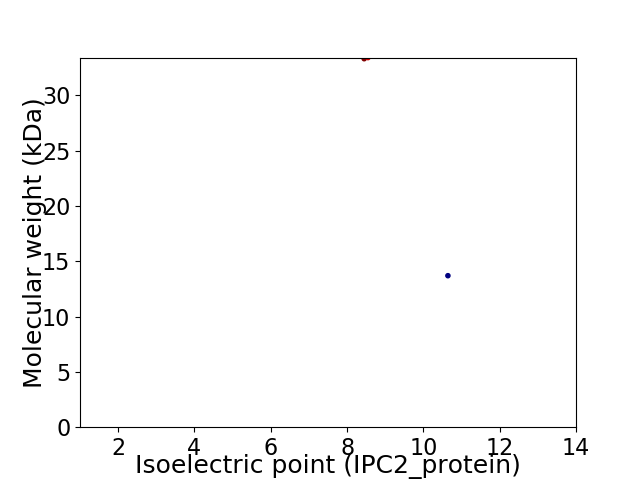
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5RNT1|W5RNT1_9CIRC ATP-dependent helicase Rep OS=Anguilla anguilla circovirus OX=1420600 PE=3 SV=1
MM1 pKa = 7.35SKK3 pKa = 10.17VSVRR7 pKa = 11.84RR8 pKa = 11.84VCFTLNNYY16 pKa = 7.58TEE18 pKa = 4.44EE19 pKa = 3.99DD20 pKa = 3.56LKK22 pKa = 11.29FIEE25 pKa = 4.36EE26 pKa = 4.27HH27 pKa = 6.01FKK29 pKa = 11.1SVSKK33 pKa = 10.44YY34 pKa = 10.42LIVGKK39 pKa = 10.41KK40 pKa = 9.6VGEE43 pKa = 4.19NGTPHH48 pKa = 5.74SQGFINLKK56 pKa = 10.34KK57 pKa = 8.79KK58 pKa = 7.2THH60 pKa = 5.84FNSVKK65 pKa = 10.21KK66 pKa = 10.32LLPRR70 pKa = 11.84AHH72 pKa = 6.6IEE74 pKa = 3.88KK75 pKa = 10.78AKK77 pKa = 11.22GNDD80 pKa = 3.4QHH82 pKa = 7.21NKK84 pKa = 10.14DD85 pKa = 3.8YY86 pKa = 11.05CSKK89 pKa = 10.51QEE91 pKa = 3.61VAFEE95 pKa = 4.23HH96 pKa = 6.65GQIQGQGKK104 pKa = 10.3RR105 pKa = 11.84NDD107 pKa = 3.81LADD110 pKa = 3.66ALATVRR116 pKa = 11.84ANGGDD121 pKa = 3.21IKK123 pKa = 11.07ALEE126 pKa = 4.7DD127 pKa = 3.61SHH129 pKa = 8.15SLVYY133 pKa = 10.52AKK135 pKa = 10.34YY136 pKa = 10.13KK137 pKa = 10.5RR138 pKa = 11.84GILACIDD145 pKa = 3.26DD146 pKa = 4.75FGWKK150 pKa = 9.81KK151 pKa = 9.84PRR153 pKa = 11.84DD154 pKa = 3.82WKK156 pKa = 10.1TEE158 pKa = 3.6VHH160 pKa = 6.0VLWGIPGCGKK170 pKa = 10.0SRR172 pKa = 11.84YY173 pKa = 9.38ARR175 pKa = 11.84EE176 pKa = 4.07QAPEE180 pKa = 3.93AYY182 pKa = 9.5YY183 pKa = 10.51KK184 pKa = 10.55PRR186 pKa = 11.84GEE188 pKa = 3.71WWDD191 pKa = 3.87GYY193 pKa = 9.27TGQEE197 pKa = 4.77DD198 pKa = 4.49VILDD202 pKa = 4.42DD203 pKa = 4.22FYY205 pKa = 11.95GWLKK209 pKa = 10.53FDD211 pKa = 4.66EE212 pKa = 4.59LLKK215 pKa = 10.71ICDD218 pKa = 3.7RR219 pKa = 11.84YY220 pKa = 10.06PYY222 pKa = 9.68RR223 pKa = 11.84VPVKK227 pKa = 10.48GSFVQFTAKK236 pKa = 10.29RR237 pKa = 11.84IYY239 pKa = 9.11ITSNVAASEE248 pKa = 4.35WYY250 pKa = 9.97HH251 pKa = 5.14FQGYY255 pKa = 10.15DD256 pKa = 3.05PTALYY261 pKa = 10.9RR262 pKa = 11.84RR263 pKa = 11.84MTSYY267 pKa = 9.43LTWNDD272 pKa = 2.68MCKK275 pKa = 10.07EE276 pKa = 3.87FRR278 pKa = 11.84PMPININYY286 pKa = 9.57
MM1 pKa = 7.35SKK3 pKa = 10.17VSVRR7 pKa = 11.84RR8 pKa = 11.84VCFTLNNYY16 pKa = 7.58TEE18 pKa = 4.44EE19 pKa = 3.99DD20 pKa = 3.56LKK22 pKa = 11.29FIEE25 pKa = 4.36EE26 pKa = 4.27HH27 pKa = 6.01FKK29 pKa = 11.1SVSKK33 pKa = 10.44YY34 pKa = 10.42LIVGKK39 pKa = 10.41KK40 pKa = 9.6VGEE43 pKa = 4.19NGTPHH48 pKa = 5.74SQGFINLKK56 pKa = 10.34KK57 pKa = 8.79KK58 pKa = 7.2THH60 pKa = 5.84FNSVKK65 pKa = 10.21KK66 pKa = 10.32LLPRR70 pKa = 11.84AHH72 pKa = 6.6IEE74 pKa = 3.88KK75 pKa = 10.78AKK77 pKa = 11.22GNDD80 pKa = 3.4QHH82 pKa = 7.21NKK84 pKa = 10.14DD85 pKa = 3.8YY86 pKa = 11.05CSKK89 pKa = 10.51QEE91 pKa = 3.61VAFEE95 pKa = 4.23HH96 pKa = 6.65GQIQGQGKK104 pKa = 10.3RR105 pKa = 11.84NDD107 pKa = 3.81LADD110 pKa = 3.66ALATVRR116 pKa = 11.84ANGGDD121 pKa = 3.21IKK123 pKa = 11.07ALEE126 pKa = 4.7DD127 pKa = 3.61SHH129 pKa = 8.15SLVYY133 pKa = 10.52AKK135 pKa = 10.34YY136 pKa = 10.13KK137 pKa = 10.5RR138 pKa = 11.84GILACIDD145 pKa = 3.26DD146 pKa = 4.75FGWKK150 pKa = 9.81KK151 pKa = 9.84PRR153 pKa = 11.84DD154 pKa = 3.82WKK156 pKa = 10.1TEE158 pKa = 3.6VHH160 pKa = 6.0VLWGIPGCGKK170 pKa = 10.0SRR172 pKa = 11.84YY173 pKa = 9.38ARR175 pKa = 11.84EE176 pKa = 4.07QAPEE180 pKa = 3.93AYY182 pKa = 9.5YY183 pKa = 10.51KK184 pKa = 10.55PRR186 pKa = 11.84GEE188 pKa = 3.71WWDD191 pKa = 3.87GYY193 pKa = 9.27TGQEE197 pKa = 4.77DD198 pKa = 4.49VILDD202 pKa = 4.42DD203 pKa = 4.22FYY205 pKa = 11.95GWLKK209 pKa = 10.53FDD211 pKa = 4.66EE212 pKa = 4.59LLKK215 pKa = 10.71ICDD218 pKa = 3.7RR219 pKa = 11.84YY220 pKa = 10.06PYY222 pKa = 9.68RR223 pKa = 11.84VPVKK227 pKa = 10.48GSFVQFTAKK236 pKa = 10.29RR237 pKa = 11.84IYY239 pKa = 9.11ITSNVAASEE248 pKa = 4.35WYY250 pKa = 9.97HH251 pKa = 5.14FQGYY255 pKa = 10.15DD256 pKa = 3.05PTALYY261 pKa = 10.9RR262 pKa = 11.84RR263 pKa = 11.84MTSYY267 pKa = 9.43LTWNDD272 pKa = 2.68MCKK275 pKa = 10.07EE276 pKa = 3.87FRR278 pKa = 11.84PMPININYY286 pKa = 9.57
Molecular weight: 33.31 kDa
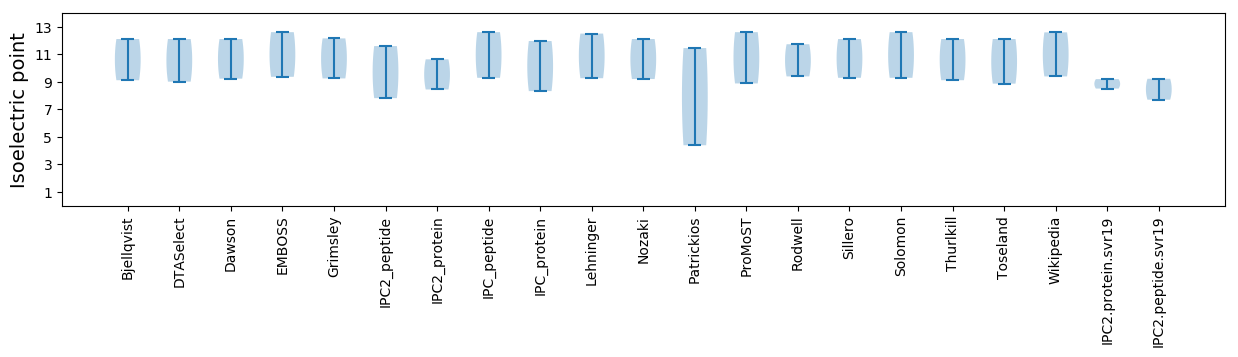
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5RNT1|W5RNT1_9CIRC ATP-dependent helicase Rep OS=Anguilla anguilla circovirus OX=1420600 PE=3 SV=1
MM1 pKa = 7.48PFYY4 pKa = 10.62RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84VYY9 pKa = 10.0RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 10.1SNRR15 pKa = 11.84RR16 pKa = 11.84PIRR19 pKa = 11.84NCQRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 9.07RR26 pKa = 11.84RR27 pKa = 11.84PIRR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 3.59RR33 pKa = 11.84NNRR36 pKa = 11.84MAVSHH41 pKa = 6.67HH42 pKa = 6.47RR43 pKa = 11.84LQIIDD48 pKa = 4.94HH49 pKa = 6.01IQMPNTANTNQTYY62 pKa = 10.87SRR64 pKa = 11.84FSRR67 pKa = 11.84FFWKK71 pKa = 9.78PSKK74 pKa = 10.25VARR77 pKa = 11.84ATPSQVPSFQAHH89 pKa = 5.86GFKK92 pKa = 10.5CVNRR96 pKa = 11.84RR97 pKa = 11.84AGPSLCSGPPGWNLRR112 pKa = 11.84SAA114 pKa = 3.93
MM1 pKa = 7.48PFYY4 pKa = 10.62RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84VYY9 pKa = 10.0RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 10.1SNRR15 pKa = 11.84RR16 pKa = 11.84PIRR19 pKa = 11.84NCQRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 9.07RR26 pKa = 11.84RR27 pKa = 11.84PIRR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 3.59RR33 pKa = 11.84NNRR36 pKa = 11.84MAVSHH41 pKa = 6.67HH42 pKa = 6.47RR43 pKa = 11.84LQIIDD48 pKa = 4.94HH49 pKa = 6.01IQMPNTANTNQTYY62 pKa = 10.87SRR64 pKa = 11.84FSRR67 pKa = 11.84FFWKK71 pKa = 9.78PSKK74 pKa = 10.25VARR77 pKa = 11.84ATPSQVPSFQAHH89 pKa = 5.86GFKK92 pKa = 10.5CVNRR96 pKa = 11.84RR97 pKa = 11.84AGPSLCSGPPGWNLRR112 pKa = 11.84SAA114 pKa = 3.93
Molecular weight: 13.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
400 |
114 |
286 |
200.0 |
23.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.0 ± 0.073 | 2.25 ± 0.198 |
4.75 ± 2.007 | 4.5 ± 1.878 |
4.75 ± 0.266 | 6.25 ± 1.421 |
3.25 ± 0.134 | 5.0 ± 0.318 |
8.25 ± 2.457 | 5.25 ± 1.357 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.75 ± 0.457 | 5.5 ± 1.241 |
5.25 ± 1.826 | 4.0 ± 0.655 |
9.75 ± 5.404 | 6.0 ± 1.437 |
4.0 ± 0.255 | 5.5 ± 0.577 |
2.5 ± 0.386 | 5.5 ± 1.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
