
Caulobacteraceae bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; unclassified Caulobacteraceae
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
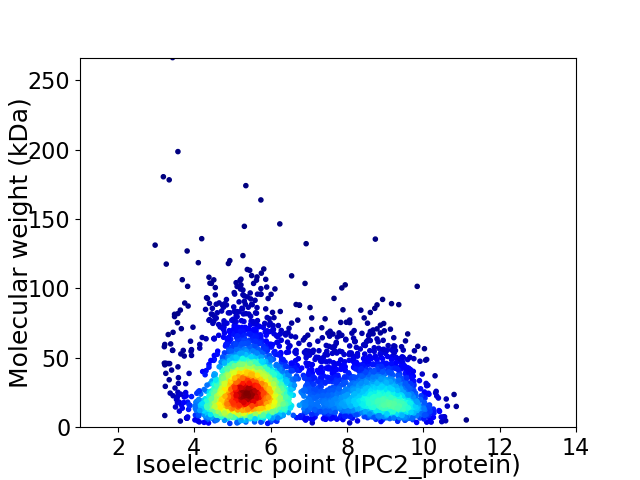
Virtual 2D-PAGE plot for 4191 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q3RUV4|A0A4Q3RUV4_9CAUL Cytochrome c4 (Fragment) OS=Caulobacteraceae bacterium OX=1978230 GN=EON95_18405 PE=4 SV=1
MM1 pKa = 7.53SGGAYY6 pKa = 9.51GQTPAPVFTGFVGPRR21 pKa = 11.84GEE23 pKa = 3.89FAKK26 pKa = 10.57RR27 pKa = 11.84RR28 pKa = 11.84FTRR31 pKa = 11.84SLGLTEE37 pKa = 4.64LCSSRR42 pKa = 11.84VWSAGATSTWCRR54 pKa = 11.84GGLTGLPRR62 pKa = 11.84HH63 pKa = 5.69TEE65 pKa = 4.07GTDD68 pKa = 3.14MPLSVLYY75 pKa = 10.74YY76 pKa = 8.69EE77 pKa = 5.58FPATGTSGDD86 pKa = 3.91DD87 pKa = 3.49FMFGYY92 pKa = 10.88ASGPGTVNTLNGGDD106 pKa = 3.67GDD108 pKa = 4.43DD109 pKa = 5.49FIMGDD114 pKa = 3.51SQNFQIAAVGNNNGSMATAQGLGDD138 pKa = 3.68SSLWSTQNNPLIADD152 pKa = 3.74PLIPYY157 pKa = 8.31TYY159 pKa = 9.76IVVEE163 pKa = 4.18ATTGQAEE170 pKa = 4.6YY171 pKa = 10.25FGTPMTAGQTITVDD185 pKa = 2.79IDD187 pKa = 3.62FGNGSPIGTGGNVDD201 pKa = 4.64LYY203 pKa = 11.71VEE205 pKa = 4.39ILDD208 pKa = 4.1SNGTVVAFSDD218 pKa = 4.81DD219 pKa = 3.32SAATDD224 pKa = 3.42GGLGSFSGLDD234 pKa = 3.31PYY236 pKa = 11.01LTFTAATSGSYY247 pKa = 10.01YY248 pKa = 9.99IRR250 pKa = 11.84VRR252 pKa = 11.84PLGEE256 pKa = 4.1TTFSASSTFILNVSMTGHH274 pKa = 7.17AVNATPIPAGADD286 pKa = 3.34TLSGGNGDD294 pKa = 4.63DD295 pKa = 5.53LIVGQDD301 pKa = 3.63GNDD304 pKa = 3.39ILHH307 pKa = 6.85GGSGMDD313 pKa = 3.82ILNGGDD319 pKa = 4.24GNDD322 pKa = 3.87TLDD325 pKa = 4.02GGTGGDD331 pKa = 4.04TLNGGAGDD339 pKa = 3.97DD340 pKa = 4.14LVWYY344 pKa = 9.99DD345 pKa = 3.82PTNFDD350 pKa = 4.44AADD353 pKa = 3.98GGTGIDD359 pKa = 3.54TLNTTRR365 pKa = 11.84YY366 pKa = 10.16ASNLVYY372 pKa = 10.88NLASGATGFGEE383 pKa = 4.29SLINFEE389 pKa = 4.64NVVTGAGNDD398 pKa = 3.8TIYY401 pKa = 10.93GSSVANRR408 pKa = 11.84IVTGGGNDD416 pKa = 3.47LVLGGDD422 pKa = 3.76GDD424 pKa = 4.56DD425 pKa = 4.52VISGGDD431 pKa = 3.7GKK433 pKa = 10.79DD434 pKa = 3.24EE435 pKa = 5.42LYY437 pKa = 11.12GQGGNDD443 pKa = 3.35TLADD447 pKa = 3.71GGGVVAEE454 pKa = 4.42TLLGGIGDD462 pKa = 3.85DD463 pKa = 3.75TYY465 pKa = 10.89IVSFRR470 pKa = 11.84EE471 pKa = 3.91SSTIEE476 pKa = 3.94YY477 pKa = 10.27ANEE480 pKa = 4.75GIDD483 pKa = 3.61TVLTTFSIFALQNNVEE499 pKa = 4.32NLTLTDD505 pKa = 3.82NGTHH509 pKa = 6.39GAGVGNALNNVITGGNGKK527 pKa = 9.89DD528 pKa = 3.52DD529 pKa = 4.01LFGRR533 pKa = 11.84DD534 pKa = 4.12GDD536 pKa = 4.12DD537 pKa = 3.48TLDD540 pKa = 4.04GNTGAANTLLGQLGNDD556 pKa = 3.53TYY558 pKa = 11.23LVRR561 pKa = 11.84VAGDD565 pKa = 3.56SVVEE569 pKa = 4.11VAGQGTDD576 pKa = 3.06TVRR579 pKa = 11.84AFVSAHH585 pKa = 4.7TLAANVEE592 pKa = 4.03NLIYY596 pKa = 10.73AGAGTFIGVGNSGGNAIYY614 pKa = 10.42GGAQADD620 pKa = 4.4DD621 pKa = 4.3LNGMGGDD628 pKa = 3.87DD629 pKa = 4.66VIIGGAGSDD638 pKa = 4.37FIQGGSGGDD647 pKa = 3.12QFRR650 pKa = 11.84YY651 pKa = 9.11MGGEE655 pKa = 3.71TGYY658 pKa = 11.15DD659 pKa = 3.37RR660 pKa = 11.84IIDD663 pKa = 3.98FVSGADD669 pKa = 3.23KK670 pKa = 11.5VMLNDD675 pKa = 4.75AFWTQTGTVDD685 pKa = 4.3FAQGAGAVATTGNSTFIHH703 pKa = 5.42NTSNGMVYY711 pKa = 10.53FDD713 pKa = 5.48LDD715 pKa = 3.75GNGAGLAVALCQLNVGQTLTASDD738 pKa = 3.97FGFFF742 pKa = 4.29
MM1 pKa = 7.53SGGAYY6 pKa = 9.51GQTPAPVFTGFVGPRR21 pKa = 11.84GEE23 pKa = 3.89FAKK26 pKa = 10.57RR27 pKa = 11.84RR28 pKa = 11.84FTRR31 pKa = 11.84SLGLTEE37 pKa = 4.64LCSSRR42 pKa = 11.84VWSAGATSTWCRR54 pKa = 11.84GGLTGLPRR62 pKa = 11.84HH63 pKa = 5.69TEE65 pKa = 4.07GTDD68 pKa = 3.14MPLSVLYY75 pKa = 10.74YY76 pKa = 8.69EE77 pKa = 5.58FPATGTSGDD86 pKa = 3.91DD87 pKa = 3.49FMFGYY92 pKa = 10.88ASGPGTVNTLNGGDD106 pKa = 3.67GDD108 pKa = 4.43DD109 pKa = 5.49FIMGDD114 pKa = 3.51SQNFQIAAVGNNNGSMATAQGLGDD138 pKa = 3.68SSLWSTQNNPLIADD152 pKa = 3.74PLIPYY157 pKa = 8.31TYY159 pKa = 9.76IVVEE163 pKa = 4.18ATTGQAEE170 pKa = 4.6YY171 pKa = 10.25FGTPMTAGQTITVDD185 pKa = 2.79IDD187 pKa = 3.62FGNGSPIGTGGNVDD201 pKa = 4.64LYY203 pKa = 11.71VEE205 pKa = 4.39ILDD208 pKa = 4.1SNGTVVAFSDD218 pKa = 4.81DD219 pKa = 3.32SAATDD224 pKa = 3.42GGLGSFSGLDD234 pKa = 3.31PYY236 pKa = 11.01LTFTAATSGSYY247 pKa = 10.01YY248 pKa = 9.99IRR250 pKa = 11.84VRR252 pKa = 11.84PLGEE256 pKa = 4.1TTFSASSTFILNVSMTGHH274 pKa = 7.17AVNATPIPAGADD286 pKa = 3.34TLSGGNGDD294 pKa = 4.63DD295 pKa = 5.53LIVGQDD301 pKa = 3.63GNDD304 pKa = 3.39ILHH307 pKa = 6.85GGSGMDD313 pKa = 3.82ILNGGDD319 pKa = 4.24GNDD322 pKa = 3.87TLDD325 pKa = 4.02GGTGGDD331 pKa = 4.04TLNGGAGDD339 pKa = 3.97DD340 pKa = 4.14LVWYY344 pKa = 9.99DD345 pKa = 3.82PTNFDD350 pKa = 4.44AADD353 pKa = 3.98GGTGIDD359 pKa = 3.54TLNTTRR365 pKa = 11.84YY366 pKa = 10.16ASNLVYY372 pKa = 10.88NLASGATGFGEE383 pKa = 4.29SLINFEE389 pKa = 4.64NVVTGAGNDD398 pKa = 3.8TIYY401 pKa = 10.93GSSVANRR408 pKa = 11.84IVTGGGNDD416 pKa = 3.47LVLGGDD422 pKa = 3.76GDD424 pKa = 4.56DD425 pKa = 4.52VISGGDD431 pKa = 3.7GKK433 pKa = 10.79DD434 pKa = 3.24EE435 pKa = 5.42LYY437 pKa = 11.12GQGGNDD443 pKa = 3.35TLADD447 pKa = 3.71GGGVVAEE454 pKa = 4.42TLLGGIGDD462 pKa = 3.85DD463 pKa = 3.75TYY465 pKa = 10.89IVSFRR470 pKa = 11.84EE471 pKa = 3.91SSTIEE476 pKa = 3.94YY477 pKa = 10.27ANEE480 pKa = 4.75GIDD483 pKa = 3.61TVLTTFSIFALQNNVEE499 pKa = 4.32NLTLTDD505 pKa = 3.82NGTHH509 pKa = 6.39GAGVGNALNNVITGGNGKK527 pKa = 9.89DD528 pKa = 3.52DD529 pKa = 4.01LFGRR533 pKa = 11.84DD534 pKa = 4.12GDD536 pKa = 4.12DD537 pKa = 3.48TLDD540 pKa = 4.04GNTGAANTLLGQLGNDD556 pKa = 3.53TYY558 pKa = 11.23LVRR561 pKa = 11.84VAGDD565 pKa = 3.56SVVEE569 pKa = 4.11VAGQGTDD576 pKa = 3.06TVRR579 pKa = 11.84AFVSAHH585 pKa = 4.7TLAANVEE592 pKa = 4.03NLIYY596 pKa = 10.73AGAGTFIGVGNSGGNAIYY614 pKa = 10.42GGAQADD620 pKa = 4.4DD621 pKa = 4.3LNGMGGDD628 pKa = 3.87DD629 pKa = 4.66VIIGGAGSDD638 pKa = 4.37FIQGGSGGDD647 pKa = 3.12QFRR650 pKa = 11.84YY651 pKa = 9.11MGGEE655 pKa = 3.71TGYY658 pKa = 11.15DD659 pKa = 3.37RR660 pKa = 11.84IIDD663 pKa = 3.98FVSGADD669 pKa = 3.23KK670 pKa = 11.5VMLNDD675 pKa = 4.75AFWTQTGTVDD685 pKa = 4.3FAQGAGAVATTGNSTFIHH703 pKa = 5.42NTSNGMVYY711 pKa = 10.53FDD713 pKa = 5.48LDD715 pKa = 3.75GNGAGLAVALCQLNVGQTLTASDD738 pKa = 3.97FGFFF742 pKa = 4.29
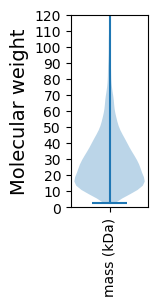
Molecular weight: 75.26 kDa
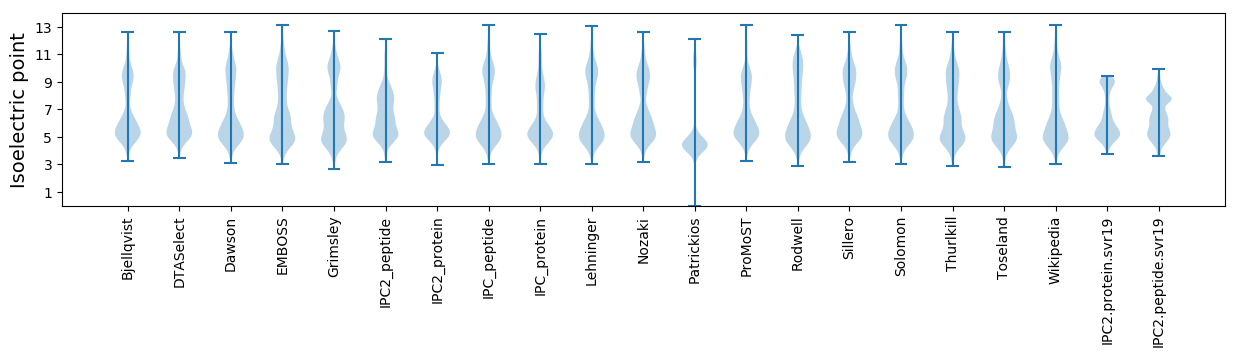
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q3S9T9|A0A4Q3S9T9_9CAUL 10 kDa chaperonin OS=Caulobacteraceae bacterium OX=1978230 GN=groS PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 5.3GYY18 pKa = 7.96RR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.84NGQKK29 pKa = 9.53IVARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 9.04KK41 pKa = 9.67LTAA44 pKa = 4.2
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 5.3GYY18 pKa = 7.96RR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.84NGQKK29 pKa = 9.53IVARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 9.04KK41 pKa = 9.67LTAA44 pKa = 4.2
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1190268 |
25 |
2727 |
284.0 |
30.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.781 ± 0.058 | 0.764 ± 0.011 |
6.016 ± 0.041 | 5.359 ± 0.038 |
3.508 ± 0.024 | 9.556 ± 0.055 |
1.735 ± 0.018 | 4.465 ± 0.028 |
3.045 ± 0.035 | 10.129 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.34 ± 0.02 | 2.359 ± 0.031 |
5.535 ± 0.035 | 3.032 ± 0.02 |
7.063 ± 0.049 | 4.963 ± 0.029 |
5.31 ± 0.043 | 7.422 ± 0.033 |
1.489 ± 0.017 | 2.128 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
