
Avon-Heathcote Estuary associated circular virus 22
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.29
Get precalculated fractions of proteins
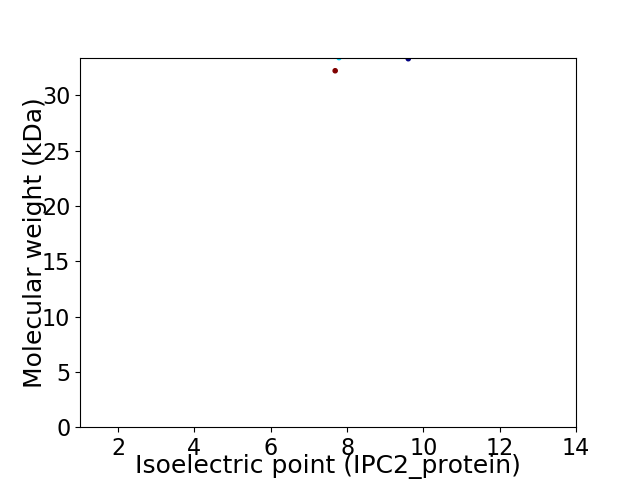
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5I9V1|A0A0C5I9V1_9CIRC Replication-associated protein OS=Avon-Heathcote Estuary associated circular virus 22 OX=1618246 PE=4 SV=1
MM1 pKa = 7.24NPIAVYY7 pKa = 10.49EE8 pKa = 4.22FTLKK12 pKa = 10.77KK13 pKa = 9.85QDD15 pKa = 3.52KK16 pKa = 9.57QNEE19 pKa = 3.99NEE21 pKa = 4.4IIKK24 pKa = 10.05ILYY27 pKa = 5.38PTIVKK32 pKa = 10.14KK33 pKa = 10.91ISFQLEE39 pKa = 4.14KK40 pKa = 10.59SDD42 pKa = 3.87EE43 pKa = 4.56GYY45 pKa = 10.29EE46 pKa = 4.14HH47 pKa = 5.79YY48 pKa = 10.16QGRR51 pKa = 11.84ISLVKK56 pKa = 9.42KK57 pKa = 10.23RR58 pKa = 11.84RR59 pKa = 11.84MKK61 pKa = 9.71EE62 pKa = 3.8TLDD65 pKa = 3.17VLKK68 pKa = 10.49PHH70 pKa = 6.82FPDD73 pKa = 2.87IHH75 pKa = 7.55ISPTCNNGLTEE86 pKa = 4.11NFYY89 pKa = 8.12TTKK92 pKa = 10.35EE93 pKa = 4.01DD94 pKa = 3.56TRR96 pKa = 11.84IDD98 pKa = 4.02GPWTEE103 pKa = 3.66KK104 pKa = 10.73SYY106 pKa = 11.42VYY108 pKa = 10.53VPRR111 pKa = 11.84QIRR114 pKa = 11.84EE115 pKa = 4.06IVDD118 pKa = 3.61LKK120 pKa = 10.27PWQKK124 pKa = 11.05SVVLLSRR131 pKa = 11.84IWDD134 pKa = 3.5TRR136 pKa = 11.84TINIIVDD143 pKa = 3.46TEE145 pKa = 4.31GNIGKK150 pKa = 8.28STLTTYY156 pKa = 10.18MGIHH160 pKa = 6.3NLAKK164 pKa = 10.23QIPFCNDD171 pKa = 2.64YY172 pKa = 11.26KK173 pKa = 11.18DD174 pKa = 4.0VLRR177 pKa = 11.84MVCDD181 pKa = 3.95MPTSGCFIIDD191 pKa = 3.41MPRR194 pKa = 11.84AIRR197 pKa = 11.84KK198 pKa = 7.17EE199 pKa = 3.94KK200 pKa = 10.14LYY202 pKa = 11.2QMYY205 pKa = 10.54SAIEE209 pKa = 4.31TIKK212 pKa = 10.67SGYY215 pKa = 10.67AYY217 pKa = 10.11DD218 pKa = 3.71EE219 pKa = 4.7RR220 pKa = 11.84YY221 pKa = 9.75HH222 pKa = 6.91FKK224 pKa = 10.73DD225 pKa = 3.53KK226 pKa = 11.49YY227 pKa = 10.66FDD229 pKa = 4.46CPCIWVFTNIIPDD242 pKa = 3.57EE243 pKa = 4.27TLLSRR248 pKa = 11.84DD249 pKa = 3.23RR250 pKa = 11.84WRR252 pKa = 11.84KK253 pKa = 7.95WEE255 pKa = 4.46IINEE259 pKa = 4.07EE260 pKa = 4.19LKK262 pKa = 11.12KK263 pKa = 10.46FDD265 pKa = 4.59PLMMRR270 pKa = 4.57
MM1 pKa = 7.24NPIAVYY7 pKa = 10.49EE8 pKa = 4.22FTLKK12 pKa = 10.77KK13 pKa = 9.85QDD15 pKa = 3.52KK16 pKa = 9.57QNEE19 pKa = 3.99NEE21 pKa = 4.4IIKK24 pKa = 10.05ILYY27 pKa = 5.38PTIVKK32 pKa = 10.14KK33 pKa = 10.91ISFQLEE39 pKa = 4.14KK40 pKa = 10.59SDD42 pKa = 3.87EE43 pKa = 4.56GYY45 pKa = 10.29EE46 pKa = 4.14HH47 pKa = 5.79YY48 pKa = 10.16QGRR51 pKa = 11.84ISLVKK56 pKa = 9.42KK57 pKa = 10.23RR58 pKa = 11.84RR59 pKa = 11.84MKK61 pKa = 9.71EE62 pKa = 3.8TLDD65 pKa = 3.17VLKK68 pKa = 10.49PHH70 pKa = 6.82FPDD73 pKa = 2.87IHH75 pKa = 7.55ISPTCNNGLTEE86 pKa = 4.11NFYY89 pKa = 8.12TTKK92 pKa = 10.35EE93 pKa = 4.01DD94 pKa = 3.56TRR96 pKa = 11.84IDD98 pKa = 4.02GPWTEE103 pKa = 3.66KK104 pKa = 10.73SYY106 pKa = 11.42VYY108 pKa = 10.53VPRR111 pKa = 11.84QIRR114 pKa = 11.84EE115 pKa = 4.06IVDD118 pKa = 3.61LKK120 pKa = 10.27PWQKK124 pKa = 11.05SVVLLSRR131 pKa = 11.84IWDD134 pKa = 3.5TRR136 pKa = 11.84TINIIVDD143 pKa = 3.46TEE145 pKa = 4.31GNIGKK150 pKa = 8.28STLTTYY156 pKa = 10.18MGIHH160 pKa = 6.3NLAKK164 pKa = 10.23QIPFCNDD171 pKa = 2.64YY172 pKa = 11.26KK173 pKa = 11.18DD174 pKa = 4.0VLRR177 pKa = 11.84MVCDD181 pKa = 3.95MPTSGCFIIDD191 pKa = 3.41MPRR194 pKa = 11.84AIRR197 pKa = 11.84KK198 pKa = 7.17EE199 pKa = 3.94KK200 pKa = 10.14LYY202 pKa = 11.2QMYY205 pKa = 10.54SAIEE209 pKa = 4.31TIKK212 pKa = 10.67SGYY215 pKa = 10.67AYY217 pKa = 10.11DD218 pKa = 3.71EE219 pKa = 4.7RR220 pKa = 11.84YY221 pKa = 9.75HH222 pKa = 6.91FKK224 pKa = 10.73DD225 pKa = 3.53KK226 pKa = 11.49YY227 pKa = 10.66FDD229 pKa = 4.46CPCIWVFTNIIPDD242 pKa = 3.57EE243 pKa = 4.27TLLSRR248 pKa = 11.84DD249 pKa = 3.23RR250 pKa = 11.84WRR252 pKa = 11.84KK253 pKa = 7.95WEE255 pKa = 4.46IINEE259 pKa = 4.07EE260 pKa = 4.19LKK262 pKa = 11.12KK263 pKa = 10.46FDD265 pKa = 4.59PLMMRR270 pKa = 4.57
Molecular weight: 32.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5I9V1|A0A0C5I9V1_9CIRC Replication-associated protein OS=Avon-Heathcote Estuary associated circular virus 22 OX=1618246 PE=4 SV=1
MM1 pKa = 7.88AFRR4 pKa = 11.84KK5 pKa = 8.64FFPRR9 pKa = 11.84KK10 pKa = 7.75KK11 pKa = 8.28TSGRR15 pKa = 11.84KK16 pKa = 7.93ATTPAKK22 pKa = 10.29RR23 pKa = 11.84RR24 pKa = 11.84TAKK27 pKa = 10.42ALPSRR32 pKa = 11.84FPSGRR37 pKa = 11.84KK38 pKa = 7.32TYY40 pKa = 9.77RR41 pKa = 11.84QTRR44 pKa = 11.84AVTRR48 pKa = 11.84VLKK51 pKa = 10.74NISEE55 pKa = 4.43TKK57 pKa = 9.97IQALTPQNAITPSPVEE73 pKa = 3.9VAPTLGPVFFTNFCLGTAPSAWVGPSGSAAFNNLNGFVWPLGSGAAQRR121 pKa = 11.84NGQYY125 pKa = 10.21IYY127 pKa = 11.02LKK129 pKa = 9.52KK130 pKa = 8.79TALNLRR136 pKa = 11.84VAMNASSRR144 pKa = 11.84HH145 pKa = 5.14GPCKK149 pKa = 10.19FRR151 pKa = 11.84VVVYY155 pKa = 9.72KK156 pKa = 10.21EE157 pKa = 3.53RR158 pKa = 11.84RR159 pKa = 11.84NLYY162 pKa = 9.41NVSGGGNPCDD172 pKa = 3.82NLFISQTGDD181 pKa = 2.99SVGFNTISPVAQRR194 pKa = 11.84TMDD197 pKa = 3.71FNTWLVNKK205 pKa = 10.2RR206 pKa = 11.84NYY208 pKa = 10.06QVVKK212 pKa = 10.77DD213 pKa = 4.02MKK215 pKa = 10.58FVLAPEE221 pKa = 4.26TLSALGSTDD230 pKa = 3.21PFNVNKK236 pKa = 10.46GYY238 pKa = 9.46TSSRR242 pKa = 11.84DD243 pKa = 3.02IRR245 pKa = 11.84LSLGHH250 pKa = 5.5FQKK253 pKa = 11.19AKK255 pKa = 10.58FGDD258 pKa = 4.18DD259 pKa = 3.64NTPEE263 pKa = 4.04DD264 pKa = 3.52QMYY267 pKa = 9.52RR268 pKa = 11.84YY269 pKa = 8.89CISVISMPMSSSEE282 pKa = 4.48APHH285 pKa = 6.66SDD287 pKa = 2.42YY288 pKa = 10.11KK289 pKa = 10.88TYY291 pKa = 10.98VNGVVSCIDD300 pKa = 3.18NN301 pKa = 3.58
MM1 pKa = 7.88AFRR4 pKa = 11.84KK5 pKa = 8.64FFPRR9 pKa = 11.84KK10 pKa = 7.75KK11 pKa = 8.28TSGRR15 pKa = 11.84KK16 pKa = 7.93ATTPAKK22 pKa = 10.29RR23 pKa = 11.84RR24 pKa = 11.84TAKK27 pKa = 10.42ALPSRR32 pKa = 11.84FPSGRR37 pKa = 11.84KK38 pKa = 7.32TYY40 pKa = 9.77RR41 pKa = 11.84QTRR44 pKa = 11.84AVTRR48 pKa = 11.84VLKK51 pKa = 10.74NISEE55 pKa = 4.43TKK57 pKa = 9.97IQALTPQNAITPSPVEE73 pKa = 3.9VAPTLGPVFFTNFCLGTAPSAWVGPSGSAAFNNLNGFVWPLGSGAAQRR121 pKa = 11.84NGQYY125 pKa = 10.21IYY127 pKa = 11.02LKK129 pKa = 9.52KK130 pKa = 8.79TALNLRR136 pKa = 11.84VAMNASSRR144 pKa = 11.84HH145 pKa = 5.14GPCKK149 pKa = 10.19FRR151 pKa = 11.84VVVYY155 pKa = 9.72KK156 pKa = 10.21EE157 pKa = 3.53RR158 pKa = 11.84RR159 pKa = 11.84NLYY162 pKa = 9.41NVSGGGNPCDD172 pKa = 3.82NLFISQTGDD181 pKa = 2.99SVGFNTISPVAQRR194 pKa = 11.84TMDD197 pKa = 3.71FNTWLVNKK205 pKa = 10.2RR206 pKa = 11.84NYY208 pKa = 10.06QVVKK212 pKa = 10.77DD213 pKa = 4.02MKK215 pKa = 10.58FVLAPEE221 pKa = 4.26TLSALGSTDD230 pKa = 3.21PFNVNKK236 pKa = 10.46GYY238 pKa = 9.46TSSRR242 pKa = 11.84DD243 pKa = 3.02IRR245 pKa = 11.84LSLGHH250 pKa = 5.5FQKK253 pKa = 11.19AKK255 pKa = 10.58FGDD258 pKa = 4.18DD259 pKa = 3.64NTPEE263 pKa = 4.04DD264 pKa = 3.52QMYY267 pKa = 9.52RR268 pKa = 11.84YY269 pKa = 8.89CISVISMPMSSSEE282 pKa = 4.48APHH285 pKa = 6.66SDD287 pKa = 2.42YY288 pKa = 10.11KK289 pKa = 10.88TYY291 pKa = 10.98VNGVVSCIDD300 pKa = 3.18NN301 pKa = 3.58
Molecular weight: 33.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
571 |
270 |
301 |
285.5 |
32.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.904 ± 1.944 | 1.926 ± 0.188 |
5.254 ± 1.136 | 4.378 ± 1.694 |
4.729 ± 0.653 | 5.254 ± 1.224 |
1.401 ± 0.287 | 6.83 ± 2.492 |
8.056 ± 1.003 | 6.305 ± 0.231 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.802 ± 0.338 | 5.954 ± 0.962 |
5.954 ± 0.49 | 3.152 ± 0.121 |
6.48 ± 0.353 | 6.83 ± 1.52 |
7.356 ± 0.203 | 6.305 ± 0.949 |
1.576 ± 0.412 | 4.553 ± 0.639 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
