
Edwardsiella phage KF-1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Kafunavirus; Edwardsiella virus KF1
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
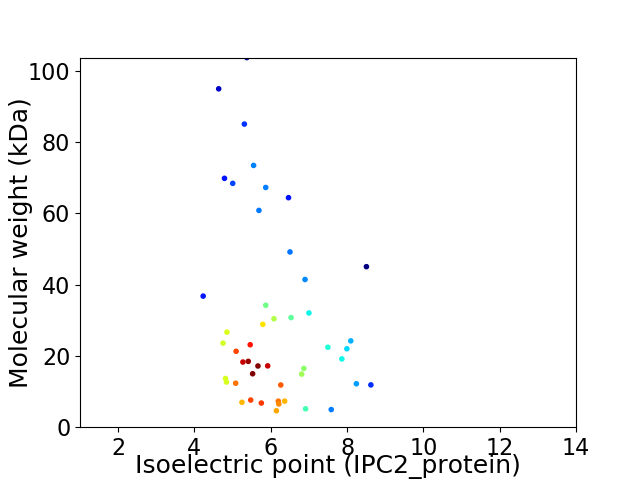
Virtual 2D-PAGE plot for 48 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4PXB9|K4PXB9_9CAUD Uncharacterized protein OS=Edwardsiella phage KF-1 OX=1244856 PE=4 SV=1
MM1 pKa = 7.21SRR3 pKa = 11.84VRR5 pKa = 11.84QIIADD10 pKa = 3.66ARR12 pKa = 11.84YY13 pKa = 10.39SLADD17 pKa = 3.53PKK19 pKa = 11.05GEE21 pKa = 3.69RR22 pKa = 11.84WTDD25 pKa = 3.16EE26 pKa = 3.68RR27 pKa = 11.84LLRR30 pKa = 11.84LLSQGQKK37 pKa = 10.84DD38 pKa = 3.58LARR41 pKa = 11.84EE42 pKa = 4.01LKK44 pKa = 10.32LLKK47 pKa = 10.68AEE49 pKa = 4.23TSLALSPGQAMYY61 pKa = 11.09KK62 pKa = 10.26LPEE65 pKa = 4.77DD66 pKa = 2.99LWLITRR72 pKa = 11.84AAFNLVRR79 pKa = 11.84IPLLSYY85 pKa = 11.08DD86 pKa = 4.32AMDD89 pKa = 4.09SADD92 pKa = 3.8AAWFARR98 pKa = 11.84TGNRR102 pKa = 11.84VEE104 pKa = 3.93NLVYY108 pKa = 10.69DD109 pKa = 4.48LRR111 pKa = 11.84NVDD114 pKa = 4.82TIRR117 pKa = 11.84VWPTPDD123 pKa = 4.31DD124 pKa = 5.96DD125 pKa = 3.93IDD127 pKa = 4.92KK128 pKa = 10.74DD129 pKa = 4.05VYY131 pKa = 10.31VFEE134 pKa = 4.89GEE136 pKa = 4.36GVLVGTMDD144 pKa = 3.53TAYY147 pKa = 10.31GVYY150 pKa = 10.55ADD152 pKa = 4.75APIDD156 pKa = 3.68KK157 pKa = 10.64VNGVLASLDD166 pKa = 3.51EE167 pKa = 4.37LASFNSVYY175 pKa = 10.79GVTADD180 pKa = 4.62LMRR183 pKa = 11.84NTLAVRR189 pKa = 11.84NTAPYY194 pKa = 9.52FGVVTEE200 pKa = 4.48FDD202 pKa = 3.73GLVPEE207 pKa = 4.66PVFGVLSDD215 pKa = 3.39IVEE218 pKa = 4.42SDD220 pKa = 4.0TIATFDD226 pKa = 3.84SLFGVATDD234 pKa = 3.66VADD237 pKa = 3.4VTGAVTIQYY246 pKa = 10.12IKK248 pKa = 11.08DD249 pKa = 3.67PDD251 pKa = 4.08DD252 pKa = 3.59VLSLDD257 pKa = 4.13GEE259 pKa = 4.61LGVPRR264 pKa = 11.84VFDD267 pKa = 3.65KK268 pKa = 11.46ALVYY272 pKa = 9.43YY273 pKa = 9.97VIGHH277 pKa = 7.22AFSDD281 pKa = 4.47DD282 pKa = 4.83LDD284 pKa = 3.72TQYY287 pKa = 10.89QQKK290 pKa = 10.3SDD292 pKa = 3.68RR293 pKa = 11.84ALSMYY298 pKa = 10.52ARR300 pKa = 11.84EE301 pKa = 4.12LTNIGEE307 pKa = 4.25QTRR310 pKa = 11.84EE311 pKa = 3.73YY312 pKa = 10.67DD313 pKa = 3.23ASATTKK319 pKa = 10.43YY320 pKa = 10.48NRR322 pKa = 11.84SSYY325 pKa = 10.64RR326 pKa = 11.84GAFDD330 pKa = 4.44DD331 pKa = 4.36
MM1 pKa = 7.21SRR3 pKa = 11.84VRR5 pKa = 11.84QIIADD10 pKa = 3.66ARR12 pKa = 11.84YY13 pKa = 10.39SLADD17 pKa = 3.53PKK19 pKa = 11.05GEE21 pKa = 3.69RR22 pKa = 11.84WTDD25 pKa = 3.16EE26 pKa = 3.68RR27 pKa = 11.84LLRR30 pKa = 11.84LLSQGQKK37 pKa = 10.84DD38 pKa = 3.58LARR41 pKa = 11.84EE42 pKa = 4.01LKK44 pKa = 10.32LLKK47 pKa = 10.68AEE49 pKa = 4.23TSLALSPGQAMYY61 pKa = 11.09KK62 pKa = 10.26LPEE65 pKa = 4.77DD66 pKa = 2.99LWLITRR72 pKa = 11.84AAFNLVRR79 pKa = 11.84IPLLSYY85 pKa = 11.08DD86 pKa = 4.32AMDD89 pKa = 4.09SADD92 pKa = 3.8AAWFARR98 pKa = 11.84TGNRR102 pKa = 11.84VEE104 pKa = 3.93NLVYY108 pKa = 10.69DD109 pKa = 4.48LRR111 pKa = 11.84NVDD114 pKa = 4.82TIRR117 pKa = 11.84VWPTPDD123 pKa = 4.31DD124 pKa = 5.96DD125 pKa = 3.93IDD127 pKa = 4.92KK128 pKa = 10.74DD129 pKa = 4.05VYY131 pKa = 10.31VFEE134 pKa = 4.89GEE136 pKa = 4.36GVLVGTMDD144 pKa = 3.53TAYY147 pKa = 10.31GVYY150 pKa = 10.55ADD152 pKa = 4.75APIDD156 pKa = 3.68KK157 pKa = 10.64VNGVLASLDD166 pKa = 3.51EE167 pKa = 4.37LASFNSVYY175 pKa = 10.79GVTADD180 pKa = 4.62LMRR183 pKa = 11.84NTLAVRR189 pKa = 11.84NTAPYY194 pKa = 9.52FGVVTEE200 pKa = 4.48FDD202 pKa = 3.73GLVPEE207 pKa = 4.66PVFGVLSDD215 pKa = 3.39IVEE218 pKa = 4.42SDD220 pKa = 4.0TIATFDD226 pKa = 3.84SLFGVATDD234 pKa = 3.66VADD237 pKa = 3.4VTGAVTIQYY246 pKa = 10.12IKK248 pKa = 11.08DD249 pKa = 3.67PDD251 pKa = 4.08DD252 pKa = 3.59VLSLDD257 pKa = 4.13GEE259 pKa = 4.61LGVPRR264 pKa = 11.84VFDD267 pKa = 3.65KK268 pKa = 11.46ALVYY272 pKa = 9.43YY273 pKa = 9.97VIGHH277 pKa = 7.22AFSDD281 pKa = 4.47DD282 pKa = 4.83LDD284 pKa = 3.72TQYY287 pKa = 10.89QQKK290 pKa = 10.3SDD292 pKa = 3.68RR293 pKa = 11.84ALSMYY298 pKa = 10.52ARR300 pKa = 11.84EE301 pKa = 4.12LTNIGEE307 pKa = 4.25QTRR310 pKa = 11.84EE311 pKa = 3.73YY312 pKa = 10.67DD313 pKa = 3.23ASATTKK319 pKa = 10.43YY320 pKa = 10.48NRR322 pKa = 11.84SSYY325 pKa = 10.64RR326 pKa = 11.84GAFDD330 pKa = 4.44DD331 pKa = 4.36
Molecular weight: 36.77 kDa
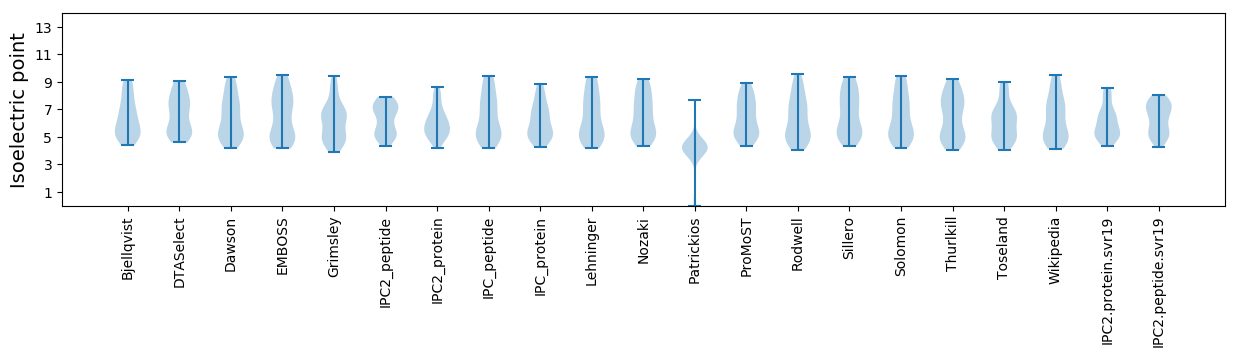
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4PWS2|K4PWS2_9CAUD Putative tailspike protein OS=Edwardsiella phage KF-1 OX=1244856 PE=4 SV=1
MM1 pKa = 7.33TPYY4 pKa = 9.4THH6 pKa = 6.68QLEE9 pKa = 4.52LSDD12 pKa = 3.57VALAILRR19 pKa = 11.84EE20 pKa = 4.02NGLAYY25 pKa = 10.47LANEE29 pKa = 3.91EE30 pKa = 4.34RR31 pKa = 11.84TGKK34 pKa = 8.06TLTAILVSEE43 pKa = 4.34KK44 pKa = 10.79SSAEE48 pKa = 3.94RR49 pKa = 11.84ILVVTKK55 pKa = 10.62KK56 pKa = 10.38KK57 pKa = 10.81ALTGWEE63 pKa = 3.87EE64 pKa = 3.47TLAAFKK70 pKa = 10.23HH71 pKa = 5.07AKK73 pKa = 9.54QYY75 pKa = 11.32VVTNYY80 pKa = 9.85HH81 pKa = 5.23QAHH84 pKa = 5.92KK85 pKa = 10.48QGSNFDD91 pKa = 4.31LVILDD96 pKa = 3.85EE97 pKa = 4.17AHH99 pKa = 6.65NYY101 pKa = 9.25ISSFPKK107 pKa = 9.16PGKK110 pKa = 8.83IHH112 pKa = 6.81KK113 pKa = 9.59EE114 pKa = 3.49LMPICKK120 pKa = 9.7GKK122 pKa = 9.62PILYY126 pKa = 10.12ISATPYY132 pKa = 10.35AQGPQMLFNQFKK144 pKa = 10.36LSSYY148 pKa = 9.62SPWYY152 pKa = 9.3KK153 pKa = 10.44HH154 pKa = 6.25SNFYY158 pKa = 10.62SWFRR162 pKa = 11.84VYY164 pKa = 10.82GKK166 pKa = 9.81PYY168 pKa = 8.36TKK170 pKa = 10.0EE171 pKa = 3.55INGINIPQYY180 pKa = 10.71DD181 pKa = 3.78RR182 pKa = 11.84CNTEE186 pKa = 3.68MVLDD190 pKa = 4.16CVKK193 pKa = 10.65HH194 pKa = 6.06LFITKK199 pKa = 8.6TRR201 pKa = 11.84ADD203 pKa = 3.64LGFEE207 pKa = 4.48HH208 pKa = 7.04EE209 pKa = 5.02PEE211 pKa = 4.98DD212 pKa = 3.48KK213 pKa = 11.15LHH215 pKa = 6.68FIEE218 pKa = 5.9LATNTKK224 pKa = 10.17AVYY227 pKa = 9.88NALLKK232 pKa = 10.95DD233 pKa = 3.6KK234 pKa = 10.58LVQLSVGKK242 pKa = 8.47VVCDD246 pKa = 3.53SAAKK250 pKa = 10.41LRR252 pKa = 11.84TTLHH256 pKa = 5.29QLEE259 pKa = 4.82GGTVKK264 pKa = 9.95IDD266 pKa = 3.59NKK268 pKa = 10.71SHH270 pKa = 5.9VLANNEE276 pKa = 3.88KK277 pKa = 8.65IDD279 pKa = 3.89YY280 pKa = 9.7IKK282 pKa = 10.94EE283 pKa = 4.11HH284 pKa = 6.58FGDD287 pKa = 3.73TGKK290 pKa = 10.54LAIMYY295 pKa = 8.62NFIGEE300 pKa = 4.48GVKK303 pKa = 10.48LNQAFKK309 pKa = 10.54HH310 pKa = 5.14AAILQATSFAEE321 pKa = 4.55GVDD324 pKa = 3.22LHH326 pKa = 7.93KK327 pKa = 10.92YY328 pKa = 10.1DD329 pKa = 3.99EE330 pKa = 4.31LVIYY334 pKa = 9.95SQDD337 pKa = 3.23YY338 pKa = 8.17STARR342 pKa = 11.84HH343 pKa = 4.32TQRR346 pKa = 11.84RR347 pKa = 11.84ARR349 pKa = 11.84QCNQKK354 pKa = 9.35RR355 pKa = 11.84DD356 pKa = 3.62KK357 pKa = 10.74PIVVHH362 pKa = 6.66FLLVANAISSQVYY375 pKa = 8.11KK376 pKa = 10.18TVSVNKK382 pKa = 9.73RR383 pKa = 11.84NYY385 pKa = 8.28VDD387 pKa = 4.29SVFEE391 pKa = 3.97RR392 pKa = 11.84TMLL395 pKa = 4.0
MM1 pKa = 7.33TPYY4 pKa = 9.4THH6 pKa = 6.68QLEE9 pKa = 4.52LSDD12 pKa = 3.57VALAILRR19 pKa = 11.84EE20 pKa = 4.02NGLAYY25 pKa = 10.47LANEE29 pKa = 3.91EE30 pKa = 4.34RR31 pKa = 11.84TGKK34 pKa = 8.06TLTAILVSEE43 pKa = 4.34KK44 pKa = 10.79SSAEE48 pKa = 3.94RR49 pKa = 11.84ILVVTKK55 pKa = 10.62KK56 pKa = 10.38KK57 pKa = 10.81ALTGWEE63 pKa = 3.87EE64 pKa = 3.47TLAAFKK70 pKa = 10.23HH71 pKa = 5.07AKK73 pKa = 9.54QYY75 pKa = 11.32VVTNYY80 pKa = 9.85HH81 pKa = 5.23QAHH84 pKa = 5.92KK85 pKa = 10.48QGSNFDD91 pKa = 4.31LVILDD96 pKa = 3.85EE97 pKa = 4.17AHH99 pKa = 6.65NYY101 pKa = 9.25ISSFPKK107 pKa = 9.16PGKK110 pKa = 8.83IHH112 pKa = 6.81KK113 pKa = 9.59EE114 pKa = 3.49LMPICKK120 pKa = 9.7GKK122 pKa = 9.62PILYY126 pKa = 10.12ISATPYY132 pKa = 10.35AQGPQMLFNQFKK144 pKa = 10.36LSSYY148 pKa = 9.62SPWYY152 pKa = 9.3KK153 pKa = 10.44HH154 pKa = 6.25SNFYY158 pKa = 10.62SWFRR162 pKa = 11.84VYY164 pKa = 10.82GKK166 pKa = 9.81PYY168 pKa = 8.36TKK170 pKa = 10.0EE171 pKa = 3.55INGINIPQYY180 pKa = 10.71DD181 pKa = 3.78RR182 pKa = 11.84CNTEE186 pKa = 3.68MVLDD190 pKa = 4.16CVKK193 pKa = 10.65HH194 pKa = 6.06LFITKK199 pKa = 8.6TRR201 pKa = 11.84ADD203 pKa = 3.64LGFEE207 pKa = 4.48HH208 pKa = 7.04EE209 pKa = 5.02PEE211 pKa = 4.98DD212 pKa = 3.48KK213 pKa = 11.15LHH215 pKa = 6.68FIEE218 pKa = 5.9LATNTKK224 pKa = 10.17AVYY227 pKa = 9.88NALLKK232 pKa = 10.95DD233 pKa = 3.6KK234 pKa = 10.58LVQLSVGKK242 pKa = 8.47VVCDD246 pKa = 3.53SAAKK250 pKa = 10.41LRR252 pKa = 11.84TTLHH256 pKa = 5.29QLEE259 pKa = 4.82GGTVKK264 pKa = 9.95IDD266 pKa = 3.59NKK268 pKa = 10.71SHH270 pKa = 5.9VLANNEE276 pKa = 3.88KK277 pKa = 8.65IDD279 pKa = 3.89YY280 pKa = 9.7IKK282 pKa = 10.94EE283 pKa = 4.11HH284 pKa = 6.58FGDD287 pKa = 3.73TGKK290 pKa = 10.54LAIMYY295 pKa = 8.62NFIGEE300 pKa = 4.48GVKK303 pKa = 10.48LNQAFKK309 pKa = 10.54HH310 pKa = 5.14AAILQATSFAEE321 pKa = 4.55GVDD324 pKa = 3.22LHH326 pKa = 7.93KK327 pKa = 10.92YY328 pKa = 10.1DD329 pKa = 3.99EE330 pKa = 4.31LVIYY334 pKa = 9.95SQDD337 pKa = 3.23YY338 pKa = 8.17STARR342 pKa = 11.84HH343 pKa = 4.32TQRR346 pKa = 11.84RR347 pKa = 11.84ARR349 pKa = 11.84QCNQKK354 pKa = 9.35RR355 pKa = 11.84DD356 pKa = 3.62KK357 pKa = 10.74PIVVHH362 pKa = 6.66FLLVANAISSQVYY375 pKa = 8.11KK376 pKa = 10.18TVSVNKK382 pKa = 9.73RR383 pKa = 11.84NYY385 pKa = 8.28VDD387 pKa = 4.29SVFEE391 pKa = 3.97RR392 pKa = 11.84TMLL395 pKa = 4.0
Molecular weight: 45.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13083 |
40 |
969 |
272.6 |
30.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.593 ± 0.527 | 1.07 ± 0.143 |
6.252 ± 0.199 | 5.916 ± 0.303 |
3.485 ± 0.21 | 7.223 ± 0.417 |
1.582 ± 0.208 | 4.701 ± 0.219 |
5.993 ± 0.286 | 8.989 ± 0.342 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.599 ± 0.187 | 4.51 ± 0.167 |
3.699 ± 0.209 | 4.257 ± 0.25 |
4.815 ± 0.242 | 6.245 ± 0.289 |
6.26 ± 0.269 | 7.536 ± 0.342 |
1.307 ± 0.15 | 3.967 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
