
Pichia sorbitophila (strain ATCC MYA-4447 / BCRC 22081 / CBS 7064 / NBRC 10061 / NRRL Y-12695) (Hybrid yeast)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Millerozyma; Millerozyma farinosa
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
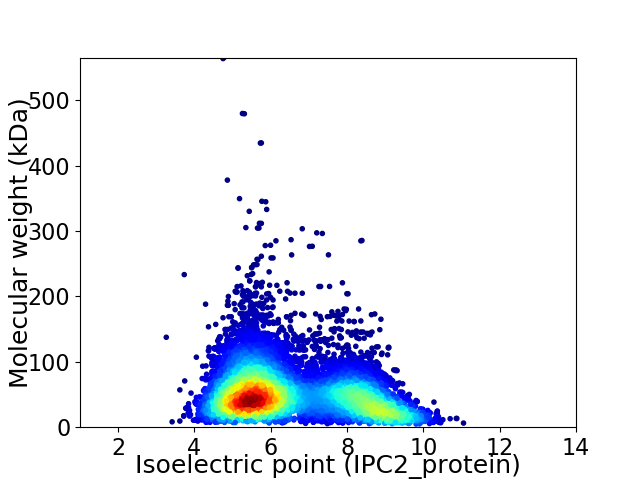
Virtual 2D-PAGE plot for 8851 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
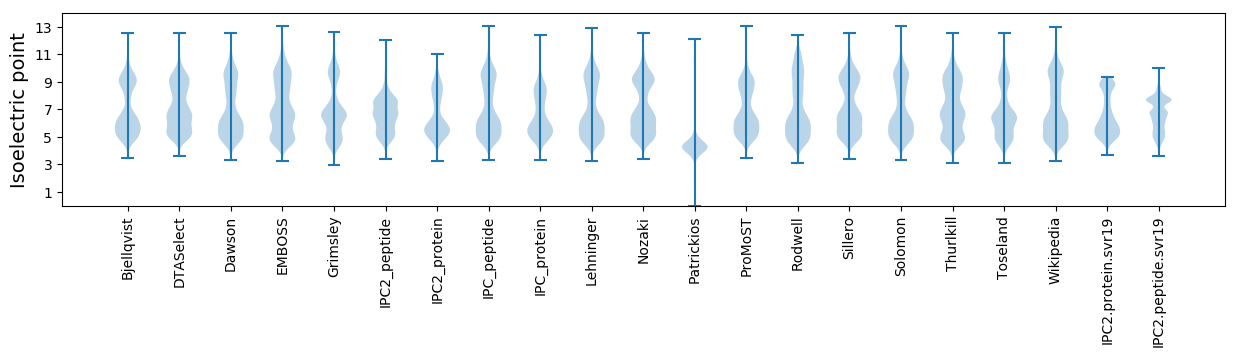
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8YQ21|G8YQ21_PICSO Piso0_000785 protein OS=Pichia sorbitophila (strain ATCC MYA-4447 / BCRC 22081 / CBS 7064 / NBRC 10061 / NRRL Y-12695) OX=559304 GN=Piso0_000785 PE=3 SV=1
MM1 pKa = 6.88VAASKK6 pKa = 11.06ALIVVVLVSTIQALLIDD23 pKa = 4.0RR24 pKa = 11.84TNDD27 pKa = 2.81IEE29 pKa = 5.11KK30 pKa = 10.35RR31 pKa = 11.84ALLSGITSLVSSVEE45 pKa = 3.99DD46 pKa = 3.61LLGGLLSAVEE56 pKa = 4.1AQLEE60 pKa = 4.54CIADD64 pKa = 3.74VVLGDD69 pKa = 4.68DD70 pKa = 4.63GQITTANEE78 pKa = 4.1DD79 pKa = 3.79EE80 pKa = 4.43FTLAVILANGFYY92 pKa = 10.3DD93 pKa = 4.04YY94 pKa = 11.66GSLDD98 pKa = 3.67SFSSLQSTLNSVADD112 pKa = 4.03NDD114 pKa = 4.89PGLLNNNTLANYY126 pKa = 9.86LGVSITDD133 pKa = 3.35NVQGVDD139 pKa = 3.34NKK141 pKa = 11.24ADD143 pKa = 3.47VLTTFIKK150 pKa = 10.66SVIEE154 pKa = 3.91VLDD157 pKa = 4.83DD158 pKa = 3.76NLSSLGGTSVTSDD171 pKa = 3.07TLANAIQEE179 pKa = 4.32KK180 pKa = 10.69AEE182 pKa = 3.86QNGIDD187 pKa = 3.77FSLSEE192 pKa = 4.58DD193 pKa = 3.47ASSDD197 pKa = 3.59SVQLVITDD205 pKa = 3.99SQWLCKK211 pKa = 10.54LLDD214 pKa = 3.72HH215 pKa = 6.89VVDD218 pKa = 4.12GLEE221 pKa = 4.02GLVDD225 pKa = 4.09CVSVTLTNLKK235 pKa = 10.02SVYY238 pKa = 10.49NLVDD242 pKa = 3.45EE243 pKa = 4.85VLGSLCPAASSSSSGVSSTAPSSTSGSSATSAPSGSSTPTSAATSGASGASSATGSGSPSGSSGTSSGSPSSSSTSGSGSPSGASSATGSGSPSGSSGTSSGSPSSSSTSGSGSPSGASSATSSGSPSGSSSATGSGSPSGSGTSGSPSGSGTSGSPSGSSASGASSVTSSGSPSGSSASGSGSSGASSATSSGSPSGASSATGSGASGSGSSSATASGSSSGSGASSVASASATGSGGSGSSGIIGDD491 pKa = 3.84LVGSIEE497 pKa = 4.01GLLGGLLSAVEE508 pKa = 4.1AQLEE512 pKa = 4.54CIADD516 pKa = 3.74VVLGDD521 pKa = 4.68DD522 pKa = 4.63GQITTANEE530 pKa = 4.1DD531 pKa = 3.79EE532 pKa = 4.43FTLAVILANGFYY544 pKa = 10.3DD545 pKa = 4.04YY546 pKa = 11.66GSLDD550 pKa = 3.67SFSSLQSTLNSVADD564 pKa = 4.03NDD566 pKa = 4.89PGLLNNNTLANYY578 pKa = 9.86LGVSITDD585 pKa = 3.35NVQGVDD591 pKa = 3.34NKK593 pKa = 11.24ADD595 pKa = 3.47VLTTFIKK602 pKa = 10.68SVIEE606 pKa = 3.85VLADD610 pKa = 3.49NLSSLGGTSVTSDD623 pKa = 3.07TLANAIQEE631 pKa = 4.32KK632 pKa = 10.69AEE634 pKa = 3.86QNGIDD639 pKa = 3.77FSLSEE644 pKa = 4.58DD645 pKa = 3.47ASSDD649 pKa = 3.59SVQLVITDD657 pKa = 3.99SQWLCKK663 pKa = 10.2LLNHH667 pKa = 5.99VVDD670 pKa = 4.25GLEE673 pKa = 3.96GLVDD677 pKa = 4.09CVSVTLTNLKK687 pKa = 10.02SVYY690 pKa = 10.49NLVDD694 pKa = 3.45EE695 pKa = 4.85VLGSLCPAASSSSSGVSSTAPSSTSGSSATSTPSGSSAPTSAATSGASGASSATGSGTPSGSSTSGSGSSSGASSATSSGSPSGSSASGSGASSATGSGTPSGSSASGSGSSSATGSGSPSGSSGTTSGSPSGSSSATGSGSPSGSSASGSGSASGASSATGSGASGSGSSSATGSGSPSGSGASGSGASSATGSGSPSSSSASGASSATSSGASSASSATGSPSGSGSSSATASGSSSGSGTSSVASASATGSGGSGSSGIIGDD960 pKa = 3.84LVGSIEE966 pKa = 4.13GLLSGLLSAVEE977 pKa = 4.51SKK979 pKa = 10.93LEE981 pKa = 4.38CIADD985 pKa = 3.68VVLGDD990 pKa = 4.15NPQSTSATEE999 pKa = 3.83DD1000 pKa = 3.54EE1001 pKa = 5.0YY1002 pKa = 11.24ILAVILANGFYY1013 pKa = 10.28DD1014 pKa = 4.16YY1015 pKa = 11.21RR1016 pKa = 11.84SLDD1019 pKa = 3.59SFDD1022 pKa = 4.64SLQSTLNSVAEE1033 pKa = 4.19NDD1035 pKa = 4.2PDD1037 pKa = 4.05LLNNNTLANYY1047 pKa = 9.86LGVSITDD1054 pKa = 3.35NVQGVDD1060 pKa = 3.34NKK1062 pKa = 11.24ADD1064 pKa = 3.47VLTTFIKK1071 pKa = 10.68SVIEE1075 pKa = 3.85VLADD1079 pKa = 3.49NLSSLGGTSVTSDD1092 pKa = 3.07TLANAIQEE1100 pKa = 4.32KK1101 pKa = 10.69AEE1103 pKa = 3.86QNGIDD1108 pKa = 3.77FSLSEE1113 pKa = 4.58DD1114 pKa = 3.47ASSDD1118 pKa = 3.59SVQLVITDD1126 pKa = 3.99SQWLCKK1132 pKa = 10.2LLNHH1136 pKa = 5.99VVDD1139 pKa = 4.16GLEE1142 pKa = 4.14GLVQCTVVDD1151 pKa = 4.56LFDD1154 pKa = 4.52LKK1156 pKa = 10.93SVYY1159 pKa = 10.87NLVDD1163 pKa = 3.45EE1164 pKa = 4.85VLGSLCPTASSSSSAVSSTAPSSTSGSSATSAPSGSSTPTSAATSGASGASSASGSGSSGSGSPSGSGASSATSSGSTSGSGASGSGSSSATSSGSPSGSGASGASGSSSATGSATGSGSPSGSSGTSSGSPSGASSATGSGASGSGSSSATGSGSPSGSGASGSGSSSATSSGSPSGSSTSGSSASGASSATGSGSPSGTSSSSASGASSATSSGSPSGSGASGSGSSSATSSGSPSGSGASGSGSSSATSSGSSSGSGASGASGSSSATGSGSSSGSGASSATSTGSPSGSGSSGSGSSSATGSTTGSGSPSGSGASGSGSSSATSSGSPSGASSATGSGASGSGSSSATGSGSPSGSGASGSGSSSATSSGSPSGSGASSATSSGSTSGSGASGSGSSSATSSGSPSGSGASGASGSSSATGSATGSGSPSGSSGTSSGSPSGASSATGSGASGSGSSSATGSGSPSGSGASGSGSSSATSSGSPSGSSTSGSSASGASSATGSGSPSGTSSSSASGASSATSSGSPSGSGASGSGSSSATSSGSSSGSGASGASGSSSATGSGSSSGSGASSATSTGSPSGSGSSGSGSSSATGSTTGSGSPSGSGASGSGSSSATSSGSPSGSGASGASGSSSATGSATGSGSPSGSSGTSSGSPSGSSASGASSASGTGASGASGSSSATSSGSASGTGASGSGSSSATGSATGSGSASGSGASSVASASATGSGGSGSSGLIGDD1905 pKa = 4.63LVGSIEE1911 pKa = 4.13GLLSGLLSAVEE1922 pKa = 4.46SKK1924 pKa = 10.98LEE1926 pKa = 4.36SIAEE1930 pKa = 4.18VVLGDD1935 pKa = 3.49NAQISSAIEE1944 pKa = 3.74DD1945 pKa = 4.07EE1946 pKa = 4.73YY1947 pKa = 11.22ILAVILANGFYY1958 pKa = 10.28DD1959 pKa = 3.83YY1960 pKa = 10.98RR1961 pKa = 11.84SSDD1964 pKa = 3.48SFNSLQSTLNSVAEE1978 pKa = 4.19NDD1980 pKa = 4.2PDD1982 pKa = 4.05LLNNNTLANYY1992 pKa = 9.73LGVGITDD1999 pKa = 3.34NVQGVDD2005 pKa = 3.31NKK2007 pKa = 11.2ADD2009 pKa = 3.54VLATFIKK2016 pKa = 10.62SVIEE2020 pKa = 3.85VLADD2024 pKa = 3.49NLSSLGGTSVTSDD2037 pKa = 3.07TLANAIQEE2045 pKa = 4.32KK2046 pKa = 10.69AEE2048 pKa = 3.86QNGIDD2053 pKa = 3.77FSLSEE2058 pKa = 4.58DD2059 pKa = 3.47ASSDD2063 pKa = 3.64SVQLVIKK2070 pKa = 9.9DD2071 pKa = 4.0PQWICQVLDD2080 pKa = 3.72LVVDD2084 pKa = 4.13GLEE2087 pKa = 4.37GLVQYY2092 pKa = 11.11DD2093 pKa = 3.67FADD2096 pKa = 3.73SFDD2099 pKa = 4.38LKK2101 pKa = 10.2TIYY2104 pKa = 10.97NLVDD2108 pKa = 3.44EE2109 pKa = 5.07VLGSLCPAASSSSSGVSSTAPSSTSGSSATSAPSGSSAPTSAATSGASGASSATGSGTPSGSGASGSGSSSATGSGSGSSGSGSSGSGSSGSGSSSATGSGSPSGSGASGASGASSATGSGSPSGASSVAPASVSGVSKK2248 pKa = 9.52GTNPTSTKK2256 pKa = 9.63TEE2258 pKa = 3.96YY2259 pKa = 10.41SHH2261 pKa = 6.75TVVTVTSCSGHH2272 pKa = 6.09KK2273 pKa = 10.31CSEE2276 pKa = 4.38TPVTTGLTTVTKK2288 pKa = 10.58DD2289 pKa = 2.98STIYY2293 pKa = 8.01TTYY2296 pKa = 10.65CPLPTEE2302 pKa = 4.33SAAPASSVAPASVTGVSKK2320 pKa = 9.64GTNPTSTKK2328 pKa = 9.63TEE2330 pKa = 3.96YY2331 pKa = 10.41SHH2333 pKa = 6.75TVVTVTSCSGNKK2345 pKa = 9.78CSEE2348 pKa = 4.39TPVTTGLTTVTKK2360 pKa = 10.58DD2361 pKa = 2.98STIYY2365 pKa = 8.01TTYY2368 pKa = 10.65CPLPTEE2374 pKa = 4.55SSAPAPVPSSKK2385 pKa = 10.5VLEE2388 pKa = 4.45SAAPASSVAPASVTGVSKK2406 pKa = 9.64GTNPTSTKK2414 pKa = 9.63TEE2416 pKa = 3.96YY2417 pKa = 10.41SHH2419 pKa = 6.75TVVTVTSCSGNKK2431 pKa = 9.78CSEE2434 pKa = 4.39TPVTTGLTTVTKK2446 pKa = 10.58DD2447 pKa = 2.98STIYY2451 pKa = 8.01TTYY2454 pKa = 10.65CPLPTEE2460 pKa = 4.55SSAPAPVPSSKK2471 pKa = 10.28VPEE2474 pKa = 4.3SAAPASSEE2482 pKa = 4.0APASKK2487 pKa = 10.65APASEE2492 pKa = 4.84GPASQAAKK2500 pKa = 10.01SSSGPAPSKK2509 pKa = 10.98APASSEE2515 pKa = 4.1APTSQAAEE2523 pKa = 4.19SSSSPVAQSSSAAAPSVNSVSEE2545 pKa = 3.93AGAIKK2550 pKa = 10.42AGISMGPLFAGAVLLLLL2567 pKa = 4.63
MM1 pKa = 6.88VAASKK6 pKa = 11.06ALIVVVLVSTIQALLIDD23 pKa = 4.0RR24 pKa = 11.84TNDD27 pKa = 2.81IEE29 pKa = 5.11KK30 pKa = 10.35RR31 pKa = 11.84ALLSGITSLVSSVEE45 pKa = 3.99DD46 pKa = 3.61LLGGLLSAVEE56 pKa = 4.1AQLEE60 pKa = 4.54CIADD64 pKa = 3.74VVLGDD69 pKa = 4.68DD70 pKa = 4.63GQITTANEE78 pKa = 4.1DD79 pKa = 3.79EE80 pKa = 4.43FTLAVILANGFYY92 pKa = 10.3DD93 pKa = 4.04YY94 pKa = 11.66GSLDD98 pKa = 3.67SFSSLQSTLNSVADD112 pKa = 4.03NDD114 pKa = 4.89PGLLNNNTLANYY126 pKa = 9.86LGVSITDD133 pKa = 3.35NVQGVDD139 pKa = 3.34NKK141 pKa = 11.24ADD143 pKa = 3.47VLTTFIKK150 pKa = 10.66SVIEE154 pKa = 3.91VLDD157 pKa = 4.83DD158 pKa = 3.76NLSSLGGTSVTSDD171 pKa = 3.07TLANAIQEE179 pKa = 4.32KK180 pKa = 10.69AEE182 pKa = 3.86QNGIDD187 pKa = 3.77FSLSEE192 pKa = 4.58DD193 pKa = 3.47ASSDD197 pKa = 3.59SVQLVITDD205 pKa = 3.99SQWLCKK211 pKa = 10.54LLDD214 pKa = 3.72HH215 pKa = 6.89VVDD218 pKa = 4.12GLEE221 pKa = 4.02GLVDD225 pKa = 4.09CVSVTLTNLKK235 pKa = 10.02SVYY238 pKa = 10.49NLVDD242 pKa = 3.45EE243 pKa = 4.85VLGSLCPAASSSSSGVSSTAPSSTSGSSATSAPSGSSTPTSAATSGASGASSATGSGSPSGSSGTSSGSPSSSSTSGSGSPSGASSATGSGSPSGSSGTSSGSPSSSSTSGSGSPSGASSATSSGSPSGSSSATGSGSPSGSGTSGSPSGSGTSGSPSGSSASGASSVTSSGSPSGSSASGSGSSGASSATSSGSPSGASSATGSGASGSGSSSATASGSSSGSGASSVASASATGSGGSGSSGIIGDD491 pKa = 3.84LVGSIEE497 pKa = 4.01GLLGGLLSAVEE508 pKa = 4.1AQLEE512 pKa = 4.54CIADD516 pKa = 3.74VVLGDD521 pKa = 4.68DD522 pKa = 4.63GQITTANEE530 pKa = 4.1DD531 pKa = 3.79EE532 pKa = 4.43FTLAVILANGFYY544 pKa = 10.3DD545 pKa = 4.04YY546 pKa = 11.66GSLDD550 pKa = 3.67SFSSLQSTLNSVADD564 pKa = 4.03NDD566 pKa = 4.89PGLLNNNTLANYY578 pKa = 9.86LGVSITDD585 pKa = 3.35NVQGVDD591 pKa = 3.34NKK593 pKa = 11.24ADD595 pKa = 3.47VLTTFIKK602 pKa = 10.68SVIEE606 pKa = 3.85VLADD610 pKa = 3.49NLSSLGGTSVTSDD623 pKa = 3.07TLANAIQEE631 pKa = 4.32KK632 pKa = 10.69AEE634 pKa = 3.86QNGIDD639 pKa = 3.77FSLSEE644 pKa = 4.58DD645 pKa = 3.47ASSDD649 pKa = 3.59SVQLVITDD657 pKa = 3.99SQWLCKK663 pKa = 10.2LLNHH667 pKa = 5.99VVDD670 pKa = 4.25GLEE673 pKa = 3.96GLVDD677 pKa = 4.09CVSVTLTNLKK687 pKa = 10.02SVYY690 pKa = 10.49NLVDD694 pKa = 3.45EE695 pKa = 4.85VLGSLCPAASSSSSGVSSTAPSSTSGSSATSTPSGSSAPTSAATSGASGASSATGSGTPSGSSTSGSGSSSGASSATSSGSPSGSSASGSGASSATGSGTPSGSSASGSGSSSATGSGSPSGSSGTTSGSPSGSSSATGSGSPSGSSASGSGSASGASSATGSGASGSGSSSATGSGSPSGSGASGSGASSATGSGSPSSSSASGASSATSSGASSASSATGSPSGSGSSSATASGSSSGSGTSSVASASATGSGGSGSSGIIGDD960 pKa = 3.84LVGSIEE966 pKa = 4.13GLLSGLLSAVEE977 pKa = 4.51SKK979 pKa = 10.93LEE981 pKa = 4.38CIADD985 pKa = 3.68VVLGDD990 pKa = 4.15NPQSTSATEE999 pKa = 3.83DD1000 pKa = 3.54EE1001 pKa = 5.0YY1002 pKa = 11.24ILAVILANGFYY1013 pKa = 10.28DD1014 pKa = 4.16YY1015 pKa = 11.21RR1016 pKa = 11.84SLDD1019 pKa = 3.59SFDD1022 pKa = 4.64SLQSTLNSVAEE1033 pKa = 4.19NDD1035 pKa = 4.2PDD1037 pKa = 4.05LLNNNTLANYY1047 pKa = 9.86LGVSITDD1054 pKa = 3.35NVQGVDD1060 pKa = 3.34NKK1062 pKa = 11.24ADD1064 pKa = 3.47VLTTFIKK1071 pKa = 10.68SVIEE1075 pKa = 3.85VLADD1079 pKa = 3.49NLSSLGGTSVTSDD1092 pKa = 3.07TLANAIQEE1100 pKa = 4.32KK1101 pKa = 10.69AEE1103 pKa = 3.86QNGIDD1108 pKa = 3.77FSLSEE1113 pKa = 4.58DD1114 pKa = 3.47ASSDD1118 pKa = 3.59SVQLVITDD1126 pKa = 3.99SQWLCKK1132 pKa = 10.2LLNHH1136 pKa = 5.99VVDD1139 pKa = 4.16GLEE1142 pKa = 4.14GLVQCTVVDD1151 pKa = 4.56LFDD1154 pKa = 4.52LKK1156 pKa = 10.93SVYY1159 pKa = 10.87NLVDD1163 pKa = 3.45EE1164 pKa = 4.85VLGSLCPTASSSSSAVSSTAPSSTSGSSATSAPSGSSTPTSAATSGASGASSASGSGSSGSGSPSGSGASSATSSGSTSGSGASGSGSSSATSSGSPSGSGASGASGSSSATGSATGSGSPSGSSGTSSGSPSGASSATGSGASGSGSSSATGSGSPSGSGASGSGSSSATSSGSPSGSSTSGSSASGASSATGSGSPSGTSSSSASGASSATSSGSPSGSGASGSGSSSATSSGSPSGSGASGSGSSSATSSGSSSGSGASGASGSSSATGSGSSSGSGASSATSTGSPSGSGSSGSGSSSATGSTTGSGSPSGSGASGSGSSSATSSGSPSGASSATGSGASGSGSSSATGSGSPSGSGASGSGSSSATSSGSPSGSGASSATSSGSTSGSGASGSGSSSATSSGSPSGSGASGASGSSSATGSATGSGSPSGSSGTSSGSPSGASSATGSGASGSGSSSATGSGSPSGSGASGSGSSSATSSGSPSGSSTSGSSASGASSATGSGSPSGTSSSSASGASSATSSGSPSGSGASGSGSSSATSSGSSSGSGASGASGSSSATGSGSSSGSGASSATSTGSPSGSGSSGSGSSSATGSTTGSGSPSGSGASGSGSSSATSSGSPSGSGASGASGSSSATGSATGSGSPSGSSGTSSGSPSGSSASGASSASGTGASGASGSSSATSSGSASGTGASGSGSSSATGSATGSGSASGSGASSVASASATGSGGSGSSGLIGDD1905 pKa = 4.63LVGSIEE1911 pKa = 4.13GLLSGLLSAVEE1922 pKa = 4.46SKK1924 pKa = 10.98LEE1926 pKa = 4.36SIAEE1930 pKa = 4.18VVLGDD1935 pKa = 3.49NAQISSAIEE1944 pKa = 3.74DD1945 pKa = 4.07EE1946 pKa = 4.73YY1947 pKa = 11.22ILAVILANGFYY1958 pKa = 10.28DD1959 pKa = 3.83YY1960 pKa = 10.98RR1961 pKa = 11.84SSDD1964 pKa = 3.48SFNSLQSTLNSVAEE1978 pKa = 4.19NDD1980 pKa = 4.2PDD1982 pKa = 4.05LLNNNTLANYY1992 pKa = 9.73LGVGITDD1999 pKa = 3.34NVQGVDD2005 pKa = 3.31NKK2007 pKa = 11.2ADD2009 pKa = 3.54VLATFIKK2016 pKa = 10.62SVIEE2020 pKa = 3.85VLADD2024 pKa = 3.49NLSSLGGTSVTSDD2037 pKa = 3.07TLANAIQEE2045 pKa = 4.32KK2046 pKa = 10.69AEE2048 pKa = 3.86QNGIDD2053 pKa = 3.77FSLSEE2058 pKa = 4.58DD2059 pKa = 3.47ASSDD2063 pKa = 3.64SVQLVIKK2070 pKa = 9.9DD2071 pKa = 4.0PQWICQVLDD2080 pKa = 3.72LVVDD2084 pKa = 4.13GLEE2087 pKa = 4.37GLVQYY2092 pKa = 11.11DD2093 pKa = 3.67FADD2096 pKa = 3.73SFDD2099 pKa = 4.38LKK2101 pKa = 10.2TIYY2104 pKa = 10.97NLVDD2108 pKa = 3.44EE2109 pKa = 5.07VLGSLCPAASSSSSGVSSTAPSSTSGSSATSAPSGSSAPTSAATSGASGASSATGSGTPSGSGASGSGSSSATGSGSGSSGSGSSGSGSSGSGSSSATGSGSPSGSGASGASGASSATGSGSPSGASSVAPASVSGVSKK2248 pKa = 9.52GTNPTSTKK2256 pKa = 9.63TEE2258 pKa = 3.96YY2259 pKa = 10.41SHH2261 pKa = 6.75TVVTVTSCSGHH2272 pKa = 6.09KK2273 pKa = 10.31CSEE2276 pKa = 4.38TPVTTGLTTVTKK2288 pKa = 10.58DD2289 pKa = 2.98STIYY2293 pKa = 8.01TTYY2296 pKa = 10.65CPLPTEE2302 pKa = 4.33SAAPASSVAPASVTGVSKK2320 pKa = 9.64GTNPTSTKK2328 pKa = 9.63TEE2330 pKa = 3.96YY2331 pKa = 10.41SHH2333 pKa = 6.75TVVTVTSCSGNKK2345 pKa = 9.78CSEE2348 pKa = 4.39TPVTTGLTTVTKK2360 pKa = 10.58DD2361 pKa = 2.98STIYY2365 pKa = 8.01TTYY2368 pKa = 10.65CPLPTEE2374 pKa = 4.55SSAPAPVPSSKK2385 pKa = 10.5VLEE2388 pKa = 4.45SAAPASSVAPASVTGVSKK2406 pKa = 9.64GTNPTSTKK2414 pKa = 9.63TEE2416 pKa = 3.96YY2417 pKa = 10.41SHH2419 pKa = 6.75TVVTVTSCSGNKK2431 pKa = 9.78CSEE2434 pKa = 4.39TPVTTGLTTVTKK2446 pKa = 10.58DD2447 pKa = 2.98STIYY2451 pKa = 8.01TTYY2454 pKa = 10.65CPLPTEE2460 pKa = 4.55SSAPAPVPSSKK2471 pKa = 10.28VPEE2474 pKa = 4.3SAAPASSEE2482 pKa = 4.0APASKK2487 pKa = 10.65APASEE2492 pKa = 4.84GPASQAAKK2500 pKa = 10.01SSSGPAPSKK2509 pKa = 10.98APASSEE2515 pKa = 4.1APTSQAAEE2523 pKa = 4.19SSSSPVAQSSSAAAPSVNSVSEE2545 pKa = 3.93AGAIKK2550 pKa = 10.42AGISMGPLFAGAVLLLLL2567 pKa = 4.63
Molecular weight: 233.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8YHK2|G8YHK2_PICSO Piso0_003239 protein OS=Pichia sorbitophila (strain ATCC MYA-4447 / BCRC 22081 / CBS 7064 / NBRC 10061 / NRRL Y-12695) OX=559304 GN=Piso0_003239 PE=4 SV=1
MM1 pKa = 7.94PSQKK5 pKa = 10.35SFRR8 pKa = 11.84VKK10 pKa = 10.47QKK12 pKa = 9.98LAKK15 pKa = 9.67AQKK18 pKa = 9.21QNRR21 pKa = 11.84PLPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNKK34 pKa = 8.39IRR36 pKa = 11.84YY37 pKa = 5.83NAKK40 pKa = 8.24RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.11WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 11.12LNII51 pKa = 3.83
MM1 pKa = 7.94PSQKK5 pKa = 10.35SFRR8 pKa = 11.84VKK10 pKa = 10.47QKK12 pKa = 9.98LAKK15 pKa = 9.67AQKK18 pKa = 9.21QNRR21 pKa = 11.84PLPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNKK34 pKa = 8.39IRR36 pKa = 11.84YY37 pKa = 5.83NAKK40 pKa = 8.24RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.11WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 11.12LNII51 pKa = 3.83
Molecular weight: 6.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4358694 |
40 |
4944 |
492.5 |
55.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.676 ± 0.023 | 1.186 ± 0.009 |
6.058 ± 0.016 | 6.694 ± 0.026 |
4.42 ± 0.018 | 5.381 ± 0.025 |
2.192 ± 0.01 | 6.35 ± 0.021 |
7.29 ± 0.027 | 9.321 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.95 ± 0.01 | 5.756 ± 0.02 |
4.267 ± 0.021 | 3.641 ± 0.018 |
4.573 ± 0.016 | 9.786 ± 0.04 |
5.195 ± 0.014 | 5.729 ± 0.015 |
0.999 ± 0.007 | 3.537 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
