
Halioglobus japonicus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Halioglobus
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
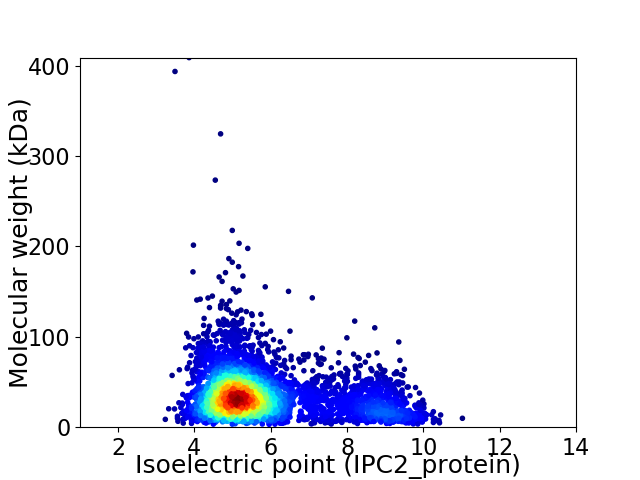
Virtual 2D-PAGE plot for 4451 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S9MWS2|A0A5S9MWS2_9GAMM Pesticin receptor OS=Halioglobus japonicus OX=930805 GN=fyuA_1 PE=3 SV=1
MM1 pKa = 7.41KK2 pKa = 10.38LVRR5 pKa = 11.84NTITLLTASLGAAMLATPAAAVPMXVASADD35 pKa = 3.45YY36 pKa = 10.5NGAVTFSLSNGLSVIGGVDD55 pKa = 3.16AQYY58 pKa = 7.92RR59 pKa = 11.84TPPGPPRR66 pKa = 11.84PYY68 pKa = 10.65QFNTSLNFGSITITPKK84 pKa = 9.54VTITTPEE91 pKa = 3.54IVLIPGSEE99 pKa = 4.0TCVPIFGCFTTPDD112 pKa = 3.29VSLPSQIIPLTPSISLTDD130 pKa = 3.6PVSVYY135 pKa = 10.7NYY137 pKa = 10.35SYY139 pKa = 7.63TTNGEE144 pKa = 4.05IPVGDD149 pKa = 3.65IFLTDD154 pKa = 4.18FGTPLLGEE162 pKa = 4.62ALTVDD167 pKa = 3.72DD168 pKa = 5.31LVRR171 pKa = 11.84EE172 pKa = 4.2QFQSGATSASEE183 pKa = 3.99SGTVVGDD190 pKa = 3.31LVGSYY195 pKa = 9.7EE196 pKa = 4.2YY197 pKa = 10.89EE198 pKa = 4.23GVLLPGGDD206 pKa = 3.92TIEE209 pKa = 4.92GEE211 pKa = 4.34YY212 pKa = 10.86LLNLSSPEE220 pKa = 3.8LLAEE224 pKa = 4.54LEE226 pKa = 4.19ASLLDD231 pKa = 5.03LINDD235 pKa = 3.63NTAEE239 pKa = 4.09IADD242 pKa = 3.82IALQALIASDD252 pKa = 3.9PCAGLGIGEE261 pKa = 4.93GICNDD266 pKa = 4.36VINGMDD272 pKa = 3.54SSEE275 pKa = 4.37LLVTVDD281 pKa = 4.34SIGEE285 pKa = 3.94FSSDD289 pKa = 3.1FSLQKK294 pKa = 10.62SVISVPVPATIPLFALGLALIGVNAKK320 pKa = 9.63RR321 pKa = 11.84RR322 pKa = 11.84RR323 pKa = 11.84AA324 pKa = 3.37
MM1 pKa = 7.41KK2 pKa = 10.38LVRR5 pKa = 11.84NTITLLTASLGAAMLATPAAAVPMXVASADD35 pKa = 3.45YY36 pKa = 10.5NGAVTFSLSNGLSVIGGVDD55 pKa = 3.16AQYY58 pKa = 7.92RR59 pKa = 11.84TPPGPPRR66 pKa = 11.84PYY68 pKa = 10.65QFNTSLNFGSITITPKK84 pKa = 9.54VTITTPEE91 pKa = 3.54IVLIPGSEE99 pKa = 4.0TCVPIFGCFTTPDD112 pKa = 3.29VSLPSQIIPLTPSISLTDD130 pKa = 3.6PVSVYY135 pKa = 10.7NYY137 pKa = 10.35SYY139 pKa = 7.63TTNGEE144 pKa = 4.05IPVGDD149 pKa = 3.65IFLTDD154 pKa = 4.18FGTPLLGEE162 pKa = 4.62ALTVDD167 pKa = 3.72DD168 pKa = 5.31LVRR171 pKa = 11.84EE172 pKa = 4.2QFQSGATSASEE183 pKa = 3.99SGTVVGDD190 pKa = 3.31LVGSYY195 pKa = 9.7EE196 pKa = 4.2YY197 pKa = 10.89EE198 pKa = 4.23GVLLPGGDD206 pKa = 3.92TIEE209 pKa = 4.92GEE211 pKa = 4.34YY212 pKa = 10.86LLNLSSPEE220 pKa = 3.8LLAEE224 pKa = 4.54LEE226 pKa = 4.19ASLLDD231 pKa = 5.03LINDD235 pKa = 3.63NTAEE239 pKa = 4.09IADD242 pKa = 3.82IALQALIASDD252 pKa = 3.9PCAGLGIGEE261 pKa = 4.93GICNDD266 pKa = 4.36VINGMDD272 pKa = 3.54SSEE275 pKa = 4.37LLVTVDD281 pKa = 4.34SIGEE285 pKa = 3.94FSSDD289 pKa = 3.1FSLQKK294 pKa = 10.62SVISVPVPATIPLFALGLALIGVNAKK320 pKa = 9.63RR321 pKa = 11.84RR322 pKa = 11.84RR323 pKa = 11.84AA324 pKa = 3.37
Molecular weight: 33.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S9QST3|A0A5S9QST3_9GAMM 50S ribosomal protein L24 OS=Halioglobus japonicus OX=930805 GN=rplX PE=3 SV=1
MM1 pKa = 7.37LWGVGIIIGLVLGLTGAGGSVFAVPLLIWIVGLEE35 pKa = 3.94PSQAIGISLGVVAIAALFGVITRR58 pKa = 11.84LRR60 pKa = 11.84SGEE63 pKa = 4.17IQWLPALVYY72 pKa = 8.85ATLGSIVAPVGQWLNQRR89 pKa = 11.84IDD91 pKa = 4.21DD92 pKa = 3.81QWLMAGFGFLVLIIALRR109 pKa = 11.84LWLQASRR116 pKa = 11.84SPQEE120 pKa = 3.72TRR122 pKa = 11.84AVRR125 pKa = 11.84ASVSAGKK132 pKa = 10.15DD133 pKa = 3.08DD134 pKa = 4.36AGAVCRR140 pKa = 11.84MINNQPFRR148 pKa = 11.84IGLPCAAGMSGGAILTGILSGLFGVGGGFIIVPTLLFLTRR188 pKa = 11.84ISIRR192 pKa = 11.84QAVATSLVVISAVGISGFSSFLLGGGNPGTGILLQVAAGSIVGMTAGVFCSRR244 pKa = 11.84FLSGPVLQKK253 pKa = 10.56VFAVMMLAIGGITVYY268 pKa = 9.73TQVLL272 pKa = 3.56
MM1 pKa = 7.37LWGVGIIIGLVLGLTGAGGSVFAVPLLIWIVGLEE35 pKa = 3.94PSQAIGISLGVVAIAALFGVITRR58 pKa = 11.84LRR60 pKa = 11.84SGEE63 pKa = 4.17IQWLPALVYY72 pKa = 8.85ATLGSIVAPVGQWLNQRR89 pKa = 11.84IDD91 pKa = 4.21DD92 pKa = 3.81QWLMAGFGFLVLIIALRR109 pKa = 11.84LWLQASRR116 pKa = 11.84SPQEE120 pKa = 3.72TRR122 pKa = 11.84AVRR125 pKa = 11.84ASVSAGKK132 pKa = 10.15DD133 pKa = 3.08DD134 pKa = 4.36AGAVCRR140 pKa = 11.84MINNQPFRR148 pKa = 11.84IGLPCAAGMSGGAILTGILSGLFGVGGGFIIVPTLLFLTRR188 pKa = 11.84ISIRR192 pKa = 11.84QAVATSLVVISAVGISGFSSFLLGGGNPGTGILLQVAAGSIVGMTAGVFCSRR244 pKa = 11.84FLSGPVLQKK253 pKa = 10.56VFAVMMLAIGGITVYY268 pKa = 9.73TQVLL272 pKa = 3.56
Molecular weight: 27.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1492785 |
29 |
4260 |
335.4 |
36.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.129 ± 0.041 | 1.102 ± 0.013 |
5.936 ± 0.029 | 6.156 ± 0.035 |
3.818 ± 0.024 | 7.692 ± 0.05 |
2.171 ± 0.018 | 5.362 ± 0.027 |
3.523 ± 0.025 | 10.376 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.493 ± 0.019 | 3.499 ± 0.025 |
4.671 ± 0.024 | 4.086 ± 0.023 |
5.747 ± 0.029 | 6.393 ± 0.032 |
5.369 ± 0.033 | 7.1 ± 0.029 |
1.417 ± 0.014 | 2.954 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
