
Agitococcus lubricus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Agitococcus
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
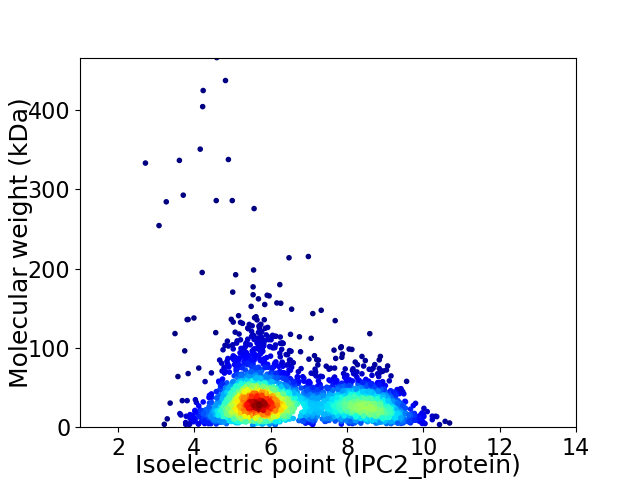
Virtual 2D-PAGE plot for 3309 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T5IVN4|A0A2T5IVN4_9GAMM Lipoyl synthase OS=Agitococcus lubricus OX=1077255 GN=lipA PE=3 SV=1
MM1 pKa = 6.92NTQIQAIFDD10 pKa = 3.84HH11 pKa = 6.99ALLANAVYY19 pKa = 10.82KK20 pKa = 10.85NLTANTTPEE29 pKa = 3.9VFAASLTGWPPALATYY45 pKa = 10.64LSTRR49 pKa = 11.84YY50 pKa = 9.88KK51 pKa = 10.67VVEE54 pKa = 4.43TYY56 pKa = 11.84NMGDD60 pKa = 3.13NVSGYY65 pKa = 10.79NGALFVGIVEE75 pKa = 4.47NDD77 pKa = 3.29NQNKK81 pKa = 10.32YY82 pKa = 10.15IFANRR87 pKa = 11.84GTEE90 pKa = 3.92PTSPSDD96 pKa = 3.97LFADD100 pKa = 4.31LLLAISGANQQLPVMQAYY118 pKa = 9.69FEE120 pKa = 4.14QALVKK125 pKa = 10.99VNGNLLTVTGHH136 pKa = 6.11SLGGYY141 pKa = 9.84LSVVLQNPDD150 pKa = 3.04EE151 pKa = 4.34NGIFIDD157 pKa = 3.82KK158 pKa = 10.18TYY160 pKa = 9.56TFNGAGIFNVALTGSIVNAGLVALGYY186 pKa = 10.6EE187 pKa = 5.05PITVSPSLPIEE198 pKa = 4.01NYY200 pKa = 8.31YY201 pKa = 9.33TVEE204 pKa = 4.01GLTVTSATGVYY215 pKa = 9.59GGVRR219 pKa = 11.84IPVNTDD225 pKa = 2.97VIGLGGVGGLEE236 pKa = 3.91GHH238 pKa = 6.81SIANLTDD245 pKa = 3.43ALAVYY250 pKa = 10.65NLMGMVDD257 pKa = 3.56SDD259 pKa = 3.46ITLAEE264 pKa = 4.11LNGIHH269 pKa = 6.42EE270 pKa = 4.95AMSKK274 pKa = 10.82DD275 pKa = 3.38GLIEE279 pKa = 3.86NDD281 pKa = 3.39ALVKK285 pKa = 10.79VFGTLLTKK293 pKa = 10.52SLTIFQQTTKK303 pKa = 11.23DD304 pKa = 4.08KK305 pKa = 11.39VDD307 pKa = 4.2LLNVSANNNNAMHH320 pKa = 6.17EE321 pKa = 4.47AIYY324 pKa = 11.3AMVNDD329 pKa = 5.15LATTAQALKK338 pKa = 10.19IIPLVSFTNQGEE350 pKa = 4.25LQLFNAQIPNNIAYY364 pKa = 9.86RR365 pKa = 11.84YY366 pKa = 8.96ALQVLNPFFVEE377 pKa = 4.57SSDD380 pKa = 3.82SANTEE385 pKa = 3.27TMYY388 pKa = 11.0QVFNEE393 pKa = 4.02HH394 pKa = 6.43QEE396 pKa = 4.0LDD398 pKa = 4.06LYY400 pKa = 11.4DD401 pKa = 5.04ADD403 pKa = 4.34TEE405 pKa = 4.38KK406 pKa = 10.83GTLTLEE412 pKa = 4.13WLEE415 pKa = 4.55KK416 pKa = 9.47RR417 pKa = 11.84HH418 pKa = 5.97EE419 pKa = 4.09MLNTQIKK426 pKa = 10.86ANIQNRR432 pKa = 11.84GFMTHH437 pKa = 5.92EE438 pKa = 4.2GEE440 pKa = 3.98YY441 pKa = 10.26NYY443 pKa = 10.82YY444 pKa = 10.21YY445 pKa = 10.59EE446 pKa = 5.91AIINNEE452 pKa = 3.74RR453 pKa = 11.84VIVKK457 pKa = 10.4DD458 pKa = 3.13MSTTSRR464 pKa = 11.84SLNDD468 pKa = 3.65PDD470 pKa = 4.01SPMVEE475 pKa = 3.81KK476 pKa = 10.08TRR478 pKa = 11.84YY479 pKa = 8.1TKK481 pKa = 10.6FADD484 pKa = 3.45KK485 pKa = 11.21NNANFTGGDD494 pKa = 3.54DD495 pKa = 3.54KK496 pKa = 11.65DD497 pKa = 3.83YY498 pKa = 11.6LFGHH502 pKa = 6.25QGNDD506 pKa = 2.84VLYY509 pKa = 10.19GLEE512 pKa = 4.11EE513 pKa = 3.87EE514 pKa = 5.51DD515 pKa = 3.76YY516 pKa = 11.63LEE518 pKa = 4.87GGEE521 pKa = 5.54GDD523 pKa = 3.74DD524 pKa = 4.18TLDD527 pKa = 3.64GGEE530 pKa = 4.39GRR532 pKa = 11.84DD533 pKa = 3.39ILVGMTGDD541 pKa = 5.03DD542 pKa = 3.71SLKK545 pKa = 10.57GDD547 pKa = 4.09KK548 pKa = 11.36GNDD551 pKa = 3.54TLEE554 pKa = 4.46GGAGKK559 pKa = 8.6DD560 pKa = 3.46TLDD563 pKa = 4.28GGEE566 pKa = 4.69DD567 pKa = 3.35HH568 pKa = 7.66DD569 pKa = 4.95KK570 pKa = 10.99LYY572 pKa = 11.21SHH574 pKa = 6.99SGDD577 pKa = 3.56DD578 pKa = 3.66TLKK581 pKa = 11.16GGLGDD586 pKa = 5.02DD587 pKa = 4.33LLDD590 pKa = 4.16GGKK593 pKa = 10.55EE594 pKa = 3.84NDD596 pKa = 4.24TLDD599 pKa = 4.26GDD601 pKa = 4.33DD602 pKa = 5.24GNDD605 pKa = 3.1ILLGQLGDD613 pKa = 5.09DD614 pKa = 3.96ILHH617 pKa = 6.28GGKK620 pKa = 10.8GNDD623 pKa = 3.38YY624 pKa = 10.58LYY626 pKa = 10.99GGEE629 pKa = 4.93GEE631 pKa = 5.2DD632 pKa = 4.06EE633 pKa = 4.55LFGEE637 pKa = 5.0AGNDD641 pKa = 3.85LIVGGKK647 pKa = 10.34GILTTAKK654 pKa = 10.27GGEE657 pKa = 4.28GNDD660 pKa = 4.54LIFSDD665 pKa = 4.59TVWAYY670 pKa = 10.11EE671 pKa = 3.8DD672 pKa = 3.82TLIGEE677 pKa = 4.45SGNDD681 pKa = 3.07YY682 pKa = 10.65LAAYY686 pKa = 9.09GGNDD690 pKa = 3.1ILKK693 pKa = 10.5GEE695 pKa = 4.54AGNDD699 pKa = 3.33YY700 pKa = 11.2LLGYY704 pKa = 8.42TGNDD708 pKa = 3.4SLLGGADD715 pKa = 3.36NDD717 pKa = 3.78VVQAGEE723 pKa = 4.21GDD725 pKa = 3.68DD726 pKa = 3.74TLVGGLHH733 pKa = 6.9HH734 pKa = 7.52DD735 pKa = 4.48LLRR738 pKa = 11.84GGNGNDD744 pKa = 3.35TYY746 pKa = 11.48SYY748 pKa = 9.18TQLEE752 pKa = 4.49GVDD755 pKa = 5.0LIEE758 pKa = 5.88DD759 pKa = 3.61SDD761 pKa = 4.14GKK763 pKa = 10.72IVIQNVTLAVTDD775 pKa = 3.84YY776 pKa = 11.64NNEE779 pKa = 3.83KK780 pKa = 10.66KK781 pKa = 10.22LWQSNTGQEE790 pKa = 3.83IRR792 pKa = 11.84RR793 pKa = 11.84LSNEE797 pKa = 3.93EE798 pKa = 4.05GNLTTVLIGQHH809 pKa = 5.63NNQIVISNFQQSQFGLTFSGGEE831 pKa = 3.82QEE833 pKa = 4.97RR834 pKa = 11.84VINVSDD840 pKa = 3.94PAKK843 pKa = 10.69QGTNQNNIIEE853 pKa = 4.54ISNHH857 pKa = 4.46VYY859 pKa = 10.41GLEE862 pKa = 4.32GNDD865 pKa = 3.45YY866 pKa = 11.2LYY868 pKa = 10.26GTVNNDD874 pKa = 2.93KK875 pKa = 10.78LYY877 pKa = 11.23GGIGGDD883 pKa = 3.71FLIGLSGDD891 pKa = 4.32DD892 pKa = 3.92YY893 pKa = 11.7IEE895 pKa = 4.78GNGDD899 pKa = 3.5DD900 pKa = 5.63DD901 pKa = 6.01IIFGGDD907 pKa = 3.53GDD909 pKa = 5.24DD910 pKa = 4.25ILYY913 pKa = 10.65GDD915 pKa = 4.81SGDD918 pKa = 4.61DD919 pKa = 3.88VIFSALKK926 pKa = 10.19IPVSFYY932 pKa = 11.1SWGLDD937 pKa = 2.97RR938 pKa = 11.84KK939 pKa = 9.97IPLSSIHH946 pKa = 5.55TQNPAEE952 pKa = 4.11GQFADD957 pKa = 3.66EE958 pKa = 5.11FDD960 pKa = 4.19NISYY964 pKa = 10.3KK965 pKa = 9.95IEE967 pKa = 3.87RR968 pKa = 11.84VLDD971 pKa = 3.89TEE973 pKa = 4.27NEE975 pKa = 4.27SYY977 pKa = 10.47TIYY980 pKa = 10.82FNGIDD985 pKa = 3.73EE986 pKa = 4.74NLDD989 pKa = 3.17VLLPVIKK996 pKa = 9.82IYY998 pKa = 11.32AEE1000 pKa = 3.85ISGEE1004 pKa = 4.01KK1005 pKa = 9.78YY1006 pKa = 9.59MLYY1009 pKa = 9.0STYY1012 pKa = 10.68SPEE1015 pKa = 4.23VYY1017 pKa = 10.35FEE1019 pKa = 4.31LTDD1022 pKa = 3.54EE1023 pKa = 4.25YY1024 pKa = 11.43NYY1026 pKa = 10.72FLDD1029 pKa = 4.34LRR1031 pKa = 11.84YY1032 pKa = 10.34EE1033 pKa = 4.16GLINNDD1039 pKa = 3.29DD1040 pKa = 4.04VYY1042 pKa = 11.5GGDD1045 pKa = 4.28GDD1047 pKa = 5.0DD1048 pKa = 4.44FLIGSGGIDD1057 pKa = 3.42KK1058 pKa = 10.63IYY1060 pKa = 10.65GNKK1063 pKa = 10.41DD1064 pKa = 2.6NDD1066 pKa = 3.55YY1067 pKa = 10.7LYY1069 pKa = 11.34GRR1071 pKa = 11.84GGDD1074 pKa = 3.55DD1075 pKa = 3.16YY1076 pKa = 11.77LYY1078 pKa = 11.11GNEE1081 pKa = 4.38GKK1083 pKa = 10.29DD1084 pKa = 3.49EE1085 pKa = 4.09IFGGDD1090 pKa = 4.17GIDD1093 pKa = 4.96LIDD1096 pKa = 5.27GGLEE1100 pKa = 3.73NDD1102 pKa = 3.89KK1103 pKa = 11.19LYY1105 pKa = 11.21GGYY1108 pKa = 10.34EE1109 pKa = 3.98SDD1111 pKa = 3.43VLYY1114 pKa = 11.23GGMGDD1119 pKa = 4.84DD1120 pKa = 4.9LVLGDD1125 pKa = 4.81FPNLYY1130 pKa = 8.86GTTTPPDD1137 pKa = 4.08SINSSRR1143 pKa = 11.84YY1144 pKa = 9.25GHH1146 pKa = 7.43DD1147 pKa = 2.54ILYY1150 pKa = 10.57GEE1152 pKa = 5.09KK1153 pKa = 11.12GNDD1156 pKa = 3.22TLFGGGADD1164 pKa = 3.76DD1165 pKa = 3.75YY1166 pKa = 11.65LYY1168 pKa = 11.28GNEE1171 pKa = 4.67DD1172 pKa = 3.86DD1173 pKa = 5.58DD1174 pKa = 4.66VVVGNEE1180 pKa = 3.8GNDD1183 pKa = 3.39YY1184 pKa = 11.29LFGGKK1189 pKa = 10.0DD1190 pKa = 3.2NDD1192 pKa = 3.43ILYY1195 pKa = 10.62GDD1197 pKa = 3.78SHH1199 pKa = 6.18EE1200 pKa = 4.19QSEE1203 pKa = 4.37AAEE1206 pKa = 4.39GNDD1209 pKa = 3.27YY1210 pKa = 11.56LEE1212 pKa = 4.73GGEE1215 pKa = 4.87GKK1217 pKa = 8.5DD1218 pKa = 3.22TLFGNGGDD1226 pKa = 3.9DD1227 pKa = 3.47VLMGGQEE1234 pKa = 3.99RR1235 pKa = 11.84DD1236 pKa = 3.39EE1237 pKa = 5.65LYY1239 pKa = 10.94DD1240 pKa = 3.84PSQLNGQLFPLKK1252 pKa = 10.63LII1254 pKa = 4.38
MM1 pKa = 6.92NTQIQAIFDD10 pKa = 3.84HH11 pKa = 6.99ALLANAVYY19 pKa = 10.82KK20 pKa = 10.85NLTANTTPEE29 pKa = 3.9VFAASLTGWPPALATYY45 pKa = 10.64LSTRR49 pKa = 11.84YY50 pKa = 9.88KK51 pKa = 10.67VVEE54 pKa = 4.43TYY56 pKa = 11.84NMGDD60 pKa = 3.13NVSGYY65 pKa = 10.79NGALFVGIVEE75 pKa = 4.47NDD77 pKa = 3.29NQNKK81 pKa = 10.32YY82 pKa = 10.15IFANRR87 pKa = 11.84GTEE90 pKa = 3.92PTSPSDD96 pKa = 3.97LFADD100 pKa = 4.31LLLAISGANQQLPVMQAYY118 pKa = 9.69FEE120 pKa = 4.14QALVKK125 pKa = 10.99VNGNLLTVTGHH136 pKa = 6.11SLGGYY141 pKa = 9.84LSVVLQNPDD150 pKa = 3.04EE151 pKa = 4.34NGIFIDD157 pKa = 3.82KK158 pKa = 10.18TYY160 pKa = 9.56TFNGAGIFNVALTGSIVNAGLVALGYY186 pKa = 10.6EE187 pKa = 5.05PITVSPSLPIEE198 pKa = 4.01NYY200 pKa = 8.31YY201 pKa = 9.33TVEE204 pKa = 4.01GLTVTSATGVYY215 pKa = 9.59GGVRR219 pKa = 11.84IPVNTDD225 pKa = 2.97VIGLGGVGGLEE236 pKa = 3.91GHH238 pKa = 6.81SIANLTDD245 pKa = 3.43ALAVYY250 pKa = 10.65NLMGMVDD257 pKa = 3.56SDD259 pKa = 3.46ITLAEE264 pKa = 4.11LNGIHH269 pKa = 6.42EE270 pKa = 4.95AMSKK274 pKa = 10.82DD275 pKa = 3.38GLIEE279 pKa = 3.86NDD281 pKa = 3.39ALVKK285 pKa = 10.79VFGTLLTKK293 pKa = 10.52SLTIFQQTTKK303 pKa = 11.23DD304 pKa = 4.08KK305 pKa = 11.39VDD307 pKa = 4.2LLNVSANNNNAMHH320 pKa = 6.17EE321 pKa = 4.47AIYY324 pKa = 11.3AMVNDD329 pKa = 5.15LATTAQALKK338 pKa = 10.19IIPLVSFTNQGEE350 pKa = 4.25LQLFNAQIPNNIAYY364 pKa = 9.86RR365 pKa = 11.84YY366 pKa = 8.96ALQVLNPFFVEE377 pKa = 4.57SSDD380 pKa = 3.82SANTEE385 pKa = 3.27TMYY388 pKa = 11.0QVFNEE393 pKa = 4.02HH394 pKa = 6.43QEE396 pKa = 4.0LDD398 pKa = 4.06LYY400 pKa = 11.4DD401 pKa = 5.04ADD403 pKa = 4.34TEE405 pKa = 4.38KK406 pKa = 10.83GTLTLEE412 pKa = 4.13WLEE415 pKa = 4.55KK416 pKa = 9.47RR417 pKa = 11.84HH418 pKa = 5.97EE419 pKa = 4.09MLNTQIKK426 pKa = 10.86ANIQNRR432 pKa = 11.84GFMTHH437 pKa = 5.92EE438 pKa = 4.2GEE440 pKa = 3.98YY441 pKa = 10.26NYY443 pKa = 10.82YY444 pKa = 10.21YY445 pKa = 10.59EE446 pKa = 5.91AIINNEE452 pKa = 3.74RR453 pKa = 11.84VIVKK457 pKa = 10.4DD458 pKa = 3.13MSTTSRR464 pKa = 11.84SLNDD468 pKa = 3.65PDD470 pKa = 4.01SPMVEE475 pKa = 3.81KK476 pKa = 10.08TRR478 pKa = 11.84YY479 pKa = 8.1TKK481 pKa = 10.6FADD484 pKa = 3.45KK485 pKa = 11.21NNANFTGGDD494 pKa = 3.54DD495 pKa = 3.54KK496 pKa = 11.65DD497 pKa = 3.83YY498 pKa = 11.6LFGHH502 pKa = 6.25QGNDD506 pKa = 2.84VLYY509 pKa = 10.19GLEE512 pKa = 4.11EE513 pKa = 3.87EE514 pKa = 5.51DD515 pKa = 3.76YY516 pKa = 11.63LEE518 pKa = 4.87GGEE521 pKa = 5.54GDD523 pKa = 3.74DD524 pKa = 4.18TLDD527 pKa = 3.64GGEE530 pKa = 4.39GRR532 pKa = 11.84DD533 pKa = 3.39ILVGMTGDD541 pKa = 5.03DD542 pKa = 3.71SLKK545 pKa = 10.57GDD547 pKa = 4.09KK548 pKa = 11.36GNDD551 pKa = 3.54TLEE554 pKa = 4.46GGAGKK559 pKa = 8.6DD560 pKa = 3.46TLDD563 pKa = 4.28GGEE566 pKa = 4.69DD567 pKa = 3.35HH568 pKa = 7.66DD569 pKa = 4.95KK570 pKa = 10.99LYY572 pKa = 11.21SHH574 pKa = 6.99SGDD577 pKa = 3.56DD578 pKa = 3.66TLKK581 pKa = 11.16GGLGDD586 pKa = 5.02DD587 pKa = 4.33LLDD590 pKa = 4.16GGKK593 pKa = 10.55EE594 pKa = 3.84NDD596 pKa = 4.24TLDD599 pKa = 4.26GDD601 pKa = 4.33DD602 pKa = 5.24GNDD605 pKa = 3.1ILLGQLGDD613 pKa = 5.09DD614 pKa = 3.96ILHH617 pKa = 6.28GGKK620 pKa = 10.8GNDD623 pKa = 3.38YY624 pKa = 10.58LYY626 pKa = 10.99GGEE629 pKa = 4.93GEE631 pKa = 5.2DD632 pKa = 4.06EE633 pKa = 4.55LFGEE637 pKa = 5.0AGNDD641 pKa = 3.85LIVGGKK647 pKa = 10.34GILTTAKK654 pKa = 10.27GGEE657 pKa = 4.28GNDD660 pKa = 4.54LIFSDD665 pKa = 4.59TVWAYY670 pKa = 10.11EE671 pKa = 3.8DD672 pKa = 3.82TLIGEE677 pKa = 4.45SGNDD681 pKa = 3.07YY682 pKa = 10.65LAAYY686 pKa = 9.09GGNDD690 pKa = 3.1ILKK693 pKa = 10.5GEE695 pKa = 4.54AGNDD699 pKa = 3.33YY700 pKa = 11.2LLGYY704 pKa = 8.42TGNDD708 pKa = 3.4SLLGGADD715 pKa = 3.36NDD717 pKa = 3.78VVQAGEE723 pKa = 4.21GDD725 pKa = 3.68DD726 pKa = 3.74TLVGGLHH733 pKa = 6.9HH734 pKa = 7.52DD735 pKa = 4.48LLRR738 pKa = 11.84GGNGNDD744 pKa = 3.35TYY746 pKa = 11.48SYY748 pKa = 9.18TQLEE752 pKa = 4.49GVDD755 pKa = 5.0LIEE758 pKa = 5.88DD759 pKa = 3.61SDD761 pKa = 4.14GKK763 pKa = 10.72IVIQNVTLAVTDD775 pKa = 3.84YY776 pKa = 11.64NNEE779 pKa = 3.83KK780 pKa = 10.66KK781 pKa = 10.22LWQSNTGQEE790 pKa = 3.83IRR792 pKa = 11.84RR793 pKa = 11.84LSNEE797 pKa = 3.93EE798 pKa = 4.05GNLTTVLIGQHH809 pKa = 5.63NNQIVISNFQQSQFGLTFSGGEE831 pKa = 3.82QEE833 pKa = 4.97RR834 pKa = 11.84VINVSDD840 pKa = 3.94PAKK843 pKa = 10.69QGTNQNNIIEE853 pKa = 4.54ISNHH857 pKa = 4.46VYY859 pKa = 10.41GLEE862 pKa = 4.32GNDD865 pKa = 3.45YY866 pKa = 11.2LYY868 pKa = 10.26GTVNNDD874 pKa = 2.93KK875 pKa = 10.78LYY877 pKa = 11.23GGIGGDD883 pKa = 3.71FLIGLSGDD891 pKa = 4.32DD892 pKa = 3.92YY893 pKa = 11.7IEE895 pKa = 4.78GNGDD899 pKa = 3.5DD900 pKa = 5.63DD901 pKa = 6.01IIFGGDD907 pKa = 3.53GDD909 pKa = 5.24DD910 pKa = 4.25ILYY913 pKa = 10.65GDD915 pKa = 4.81SGDD918 pKa = 4.61DD919 pKa = 3.88VIFSALKK926 pKa = 10.19IPVSFYY932 pKa = 11.1SWGLDD937 pKa = 2.97RR938 pKa = 11.84KK939 pKa = 9.97IPLSSIHH946 pKa = 5.55TQNPAEE952 pKa = 4.11GQFADD957 pKa = 3.66EE958 pKa = 5.11FDD960 pKa = 4.19NISYY964 pKa = 10.3KK965 pKa = 9.95IEE967 pKa = 3.87RR968 pKa = 11.84VLDD971 pKa = 3.89TEE973 pKa = 4.27NEE975 pKa = 4.27SYY977 pKa = 10.47TIYY980 pKa = 10.82FNGIDD985 pKa = 3.73EE986 pKa = 4.74NLDD989 pKa = 3.17VLLPVIKK996 pKa = 9.82IYY998 pKa = 11.32AEE1000 pKa = 3.85ISGEE1004 pKa = 4.01KK1005 pKa = 9.78YY1006 pKa = 9.59MLYY1009 pKa = 9.0STYY1012 pKa = 10.68SPEE1015 pKa = 4.23VYY1017 pKa = 10.35FEE1019 pKa = 4.31LTDD1022 pKa = 3.54EE1023 pKa = 4.25YY1024 pKa = 11.43NYY1026 pKa = 10.72FLDD1029 pKa = 4.34LRR1031 pKa = 11.84YY1032 pKa = 10.34EE1033 pKa = 4.16GLINNDD1039 pKa = 3.29DD1040 pKa = 4.04VYY1042 pKa = 11.5GGDD1045 pKa = 4.28GDD1047 pKa = 5.0DD1048 pKa = 4.44FLIGSGGIDD1057 pKa = 3.42KK1058 pKa = 10.63IYY1060 pKa = 10.65GNKK1063 pKa = 10.41DD1064 pKa = 2.6NDD1066 pKa = 3.55YY1067 pKa = 10.7LYY1069 pKa = 11.34GRR1071 pKa = 11.84GGDD1074 pKa = 3.55DD1075 pKa = 3.16YY1076 pKa = 11.77LYY1078 pKa = 11.11GNEE1081 pKa = 4.38GKK1083 pKa = 10.29DD1084 pKa = 3.49EE1085 pKa = 4.09IFGGDD1090 pKa = 4.17GIDD1093 pKa = 4.96LIDD1096 pKa = 5.27GGLEE1100 pKa = 3.73NDD1102 pKa = 3.89KK1103 pKa = 11.19LYY1105 pKa = 11.21GGYY1108 pKa = 10.34EE1109 pKa = 3.98SDD1111 pKa = 3.43VLYY1114 pKa = 11.23GGMGDD1119 pKa = 4.84DD1120 pKa = 4.9LVLGDD1125 pKa = 4.81FPNLYY1130 pKa = 8.86GTTTPPDD1137 pKa = 4.08SINSSRR1143 pKa = 11.84YY1144 pKa = 9.25GHH1146 pKa = 7.43DD1147 pKa = 2.54ILYY1150 pKa = 10.57GEE1152 pKa = 5.09KK1153 pKa = 11.12GNDD1156 pKa = 3.22TLFGGGADD1164 pKa = 3.76DD1165 pKa = 3.75YY1166 pKa = 11.65LYY1168 pKa = 11.28GNEE1171 pKa = 4.67DD1172 pKa = 3.86DD1173 pKa = 5.58DD1174 pKa = 4.66VVVGNEE1180 pKa = 3.8GNDD1183 pKa = 3.39YY1184 pKa = 11.29LFGGKK1189 pKa = 10.0DD1190 pKa = 3.2NDD1192 pKa = 3.43ILYY1195 pKa = 10.62GDD1197 pKa = 3.78SHH1199 pKa = 6.18EE1200 pKa = 4.19QSEE1203 pKa = 4.37AAEE1206 pKa = 4.39GNDD1209 pKa = 3.27YY1210 pKa = 11.56LEE1212 pKa = 4.73GGEE1215 pKa = 4.87GKK1217 pKa = 8.5DD1218 pKa = 3.22TLFGNGGDD1226 pKa = 3.9DD1227 pKa = 3.47VLMGGQEE1234 pKa = 3.99RR1235 pKa = 11.84DD1236 pKa = 3.39EE1237 pKa = 5.65LYY1239 pKa = 10.94DD1240 pKa = 3.84PSQLNGQLFPLKK1252 pKa = 10.63LII1254 pKa = 4.38
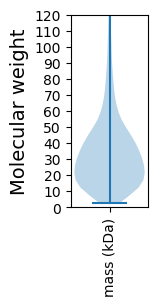
Molecular weight: 135.91 kDa
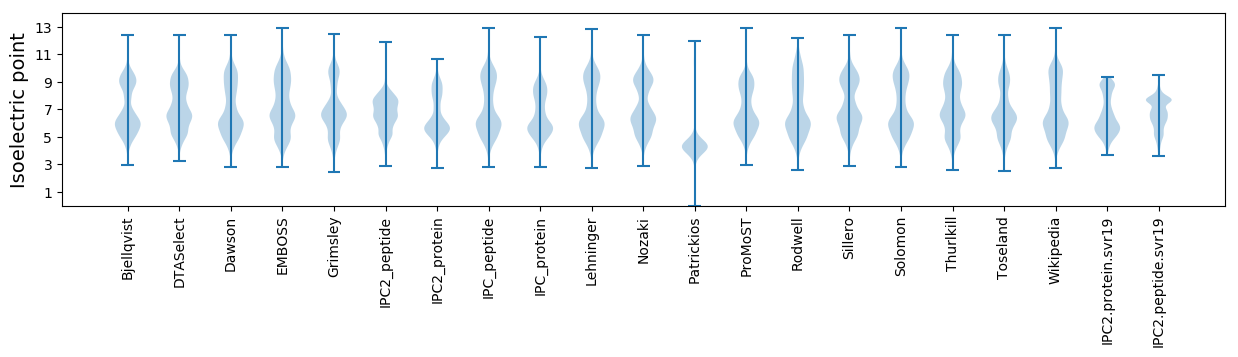
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T5J1U5|A0A2T5J1U5_9GAMM Magnesium and cobalt transporter OS=Agitococcus lubricus OX=1077255 GN=C8N29_103168 PE=4 SV=1
MM1 pKa = 7.1SAWVLFIACLVICLRR16 pKa = 11.84LFTFQRR22 pKa = 11.84KK23 pKa = 6.24GARR26 pKa = 11.84YY27 pKa = 8.87RR28 pKa = 11.84PVMSFGAYY36 pKa = 10.47LLMVFSFSIMVKK48 pKa = 10.26LALNQFPCHH57 pKa = 6.28VSPFMSLAAVGLAWLTLVNKK77 pKa = 10.42GNVAHH82 pKa = 7.07FFRR85 pKa = 11.84KK86 pKa = 9.5HH87 pKa = 3.69SHH89 pKa = 5.13GG90 pKa = 3.63
MM1 pKa = 7.1SAWVLFIACLVICLRR16 pKa = 11.84LFTFQRR22 pKa = 11.84KK23 pKa = 6.24GARR26 pKa = 11.84YY27 pKa = 8.87RR28 pKa = 11.84PVMSFGAYY36 pKa = 10.47LLMVFSFSIMVKK48 pKa = 10.26LALNQFPCHH57 pKa = 6.28VSPFMSLAAVGLAWLTLVNKK77 pKa = 10.42GNVAHH82 pKa = 7.07FFRR85 pKa = 11.84KK86 pKa = 9.5HH87 pKa = 3.69SHH89 pKa = 5.13GG90 pKa = 3.63
Molecular weight: 10.21 kDa
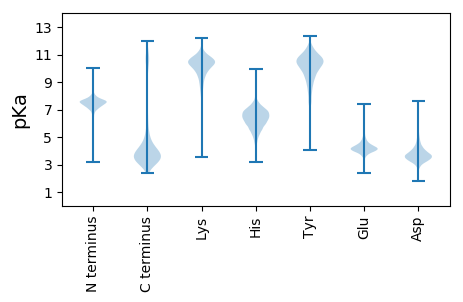
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1093339 |
26 |
4622 |
330.4 |
36.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.579 ± 0.053 | 0.987 ± 0.015 |
5.303 ± 0.04 | 5.43 ± 0.042 |
3.789 ± 0.03 | 6.646 ± 0.059 |
2.435 ± 0.027 | 6.054 ± 0.031 |
4.871 ± 0.046 | 10.919 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.368 ± 0.026 | 4.21 ± 0.059 |
4.076 ± 0.037 | 5.443 ± 0.049 |
4.898 ± 0.044 | 6.065 ± 0.041 |
5.688 ± 0.078 | 6.817 ± 0.037 |
1.332 ± 0.02 | 3.088 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
