
Synechococcus sp. (strain WH7805)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Synechococcaceae; Synechococcus; unclassified Synechococcus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
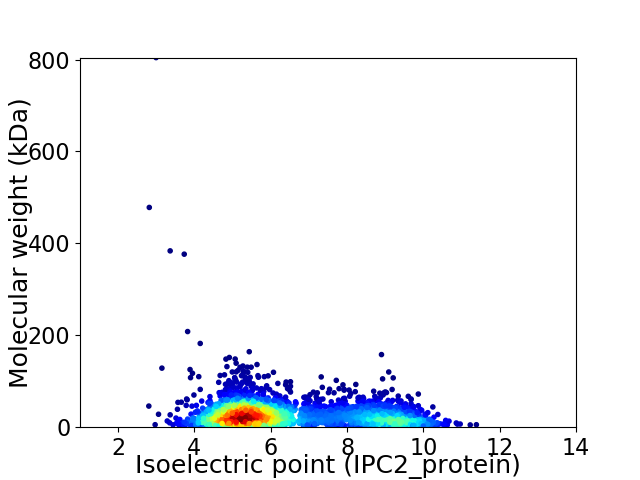
Virtual 2D-PAGE plot for 2874 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
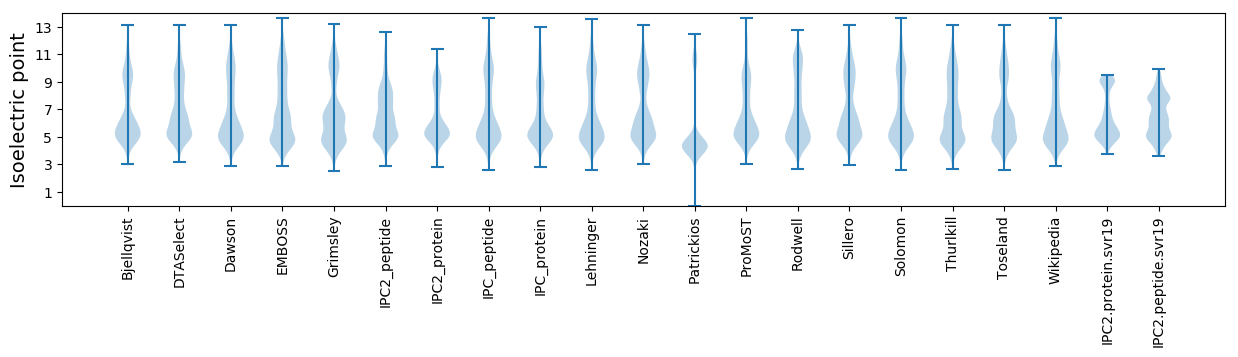
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4CWW8|A4CWW8_SYNPV Elp3 domain-containing protein OS=Synechococcus sp. (strain WH7805) OX=59931 GN=WH7805_09569 PE=4 SV=1
MM1 pKa = 7.3NLSNLIEE8 pKa = 4.32SLSAYY13 pKa = 9.62RR14 pKa = 11.84ARR16 pKa = 11.84SLHH19 pKa = 5.77TSLNQPEE26 pKa = 4.82EE27 pKa = 4.37PDD29 pKa = 3.83PDD31 pKa = 3.47ISSIDD36 pKa = 3.73NNPALTPIDD45 pKa = 3.51QPLTVARR52 pKa = 11.84GLLQNGATTPPSQQLNSTEE71 pKa = 4.28SFSEE75 pKa = 4.52AINGNNSITTEE86 pKa = 4.03EE87 pKa = 4.3EE88 pKa = 3.87ANEE91 pKa = 4.0TTNIEE96 pKa = 3.8FRR98 pKa = 11.84IEE100 pKa = 4.09SKK102 pKa = 10.51YY103 pKa = 10.99LHH105 pKa = 7.01AGRR108 pKa = 11.84CACFACSYY116 pKa = 10.94SRR118 pKa = 11.84DD119 pKa = 3.56SLEE122 pKa = 4.11TNRR125 pKa = 11.84EE126 pKa = 4.25SNSEE130 pKa = 4.1LNNTLSNADD139 pKa = 3.38PTIAFGTLNEE149 pKa = 4.15LSDD152 pKa = 4.01YY153 pKa = 9.5LTTGFWEE160 pKa = 4.19EE161 pKa = 3.61AGTYY165 pKa = 7.19TRR167 pKa = 11.84RR168 pKa = 11.84FNLGGSGLGAKK179 pKa = 9.84NGQLTYY185 pKa = 10.99NITGWADD192 pKa = 3.34DD193 pKa = 4.12SNGLSAEE200 pKa = 4.16RR201 pKa = 11.84QNLTRR206 pKa = 11.84EE207 pKa = 4.05VFNVYY212 pKa = 9.39EE213 pKa = 5.0AITGIEE219 pKa = 4.12FVEE222 pKa = 4.71VATEE226 pKa = 3.88GDD228 pKa = 4.31FRR230 pKa = 11.84FTDD233 pKa = 3.74NDD235 pKa = 3.64SGAYY239 pKa = 9.83AYY241 pKa = 10.81LGGGWYY247 pKa = 9.94DD248 pKa = 3.29QDD250 pKa = 4.71PNTGTIDD257 pKa = 3.46YY258 pKa = 9.12NAAIIDD264 pKa = 3.69YY265 pKa = 10.57SVINVASSWFYY276 pKa = 11.49GDD278 pKa = 3.68SSYY281 pKa = 11.9NSYY284 pKa = 9.32TPQTIFHH291 pKa = 7.24EE292 pKa = 4.72IGHH295 pKa = 6.51ALGLGHH301 pKa = 6.58QGQYY305 pKa = 10.39NAGNGNPTYY314 pKa = 10.38EE315 pKa = 4.43NSAQYY320 pKa = 11.53GNDD323 pKa = 2.93TWLTTMMSYY332 pKa = 9.87WSQTTNTNVNASRR345 pKa = 11.84AYY347 pKa = 10.07LQTPMTVDD355 pKa = 3.9WIALDD360 pKa = 3.81KK361 pKa = 11.2LYY363 pKa = 11.05GSQGYY368 pKa = 10.19GSFKK372 pKa = 10.64AFNGNTVYY380 pKa = 10.78GVGTNISSGTSEE392 pKa = 3.97IMNNFATMISDD403 pKa = 3.9TAYY406 pKa = 9.87TLVDD410 pKa = 3.66GSGYY414 pKa = 10.79DD415 pKa = 3.2ILNVSNYY422 pKa = 10.35SDD424 pKa = 3.48DD425 pKa = 3.68QFINLAPSEE434 pKa = 4.31LNGTLPSSSSIGGLVNNLTIGVGTILEE461 pKa = 4.17EE462 pKa = 4.95AIGGSGNDD470 pKa = 3.29TFLGNIADD478 pKa = 3.8NVFRR482 pKa = 11.84GGNGGDD488 pKa = 3.07RR489 pKa = 11.84FYY491 pKa = 11.54DD492 pKa = 3.5SFGSDD497 pKa = 2.67TYY499 pKa = 11.41YY500 pKa = 11.06GGSGTFDD507 pKa = 2.76TLYY510 pKa = 10.81FSEE513 pKa = 4.75NYY515 pKa = 10.3SDD517 pKa = 5.56FSIEE521 pKa = 4.03NLGTSLAFSRR531 pKa = 11.84NNSGLADD538 pKa = 4.04TDD540 pKa = 5.5LIWNDD545 pKa = 2.73IEE547 pKa = 5.2LVYY550 pKa = 11.06FNDD553 pKa = 4.72DD554 pKa = 3.55VIKK557 pKa = 9.85TYY559 pKa = 10.83QEE561 pKa = 3.9LLADD565 pKa = 3.95ITPINTPPTANDD577 pKa = 3.1VFVATSEE584 pKa = 4.37DD585 pKa = 4.05SNSVSGNFNANDD597 pKa = 4.43PDD599 pKa = 4.47SSDD602 pKa = 3.1QLTYY606 pKa = 10.73TITTAPTIGSVVVNSDD622 pKa = 3.12GTFTYY627 pKa = 11.06NFGNNFQSLGDD638 pKa = 3.87GDD640 pKa = 3.92STAVVFQYY648 pKa = 10.52IASDD652 pKa = 3.99DD653 pKa = 4.09NATDD657 pKa = 3.68SAPATVTITINGSNDD672 pKa = 2.91APILSGTPSALTNGTEE688 pKa = 4.67DD689 pKa = 3.05IAYY692 pKa = 10.01LLKK695 pKa = 10.96ASDD698 pKa = 4.24LLQGYY703 pKa = 9.81SDD705 pKa = 3.89IDD707 pKa = 3.94GDD709 pKa = 4.18TLSITSVTTASANGTLTPNGNGTWTYY735 pKa = 11.08TPTQDD740 pKa = 4.41LNGEE744 pKa = 4.46VVFSFVVSDD753 pKa = 4.19GNGGTANGSTSLTLDD768 pKa = 3.49PVEE771 pKa = 5.89DD772 pKa = 3.84EE773 pKa = 4.64VLLDD777 pKa = 3.78EE778 pKa = 5.44DD779 pKa = 4.05NNGIVDD785 pKa = 4.06GTEE788 pKa = 3.5LTAYY792 pKa = 10.44QLFSEE797 pKa = 5.14AGAITLKK804 pKa = 11.0NNAGQTYY811 pKa = 9.93NNSSSGNSDD820 pKa = 2.9VVAAIEE826 pKa = 4.21TDD828 pKa = 2.99NGFQVLLEE836 pKa = 4.3GTGSRR841 pKa = 11.84NDD843 pKa = 3.39TFYY846 pKa = 11.64VWNTNSNGVIIGGSGWKK863 pKa = 10.22SGDD866 pKa = 2.84IATRR870 pKa = 11.84LGWEE874 pKa = 3.9EE875 pKa = 4.07TFNFDD880 pKa = 3.69TNRR883 pKa = 11.84DD884 pKa = 3.52GTIGALILDD893 pKa = 4.83DD894 pKa = 5.09DD895 pKa = 4.67NNGIVDD901 pKa = 4.05GTEE904 pKa = 3.5LTAYY908 pKa = 10.44QLFSEE913 pKa = 5.14AGAITLKK920 pKa = 11.0NNAGQTYY927 pKa = 9.93NNSSSGNSDD936 pKa = 2.9VVAAIEE942 pKa = 4.21TDD944 pKa = 2.99NGFQVLLEE952 pKa = 4.3GTGSRR957 pKa = 11.84NDD959 pKa = 3.39TFYY962 pKa = 11.64VWNTNSNGVIIGGSGWKK979 pKa = 10.22SGDD982 pKa = 2.84IATRR986 pKa = 11.84LGWEE990 pKa = 3.9EE991 pKa = 4.07TFNFDD996 pKa = 3.69TNRR999 pKa = 11.84DD1000 pKa = 3.52GTIGALILDD1009 pKa = 4.83DD1010 pKa = 5.09DD1011 pKa = 4.67NNGIVDD1017 pKa = 4.05GTEE1020 pKa = 3.5LTAYY1024 pKa = 10.44QLFSEE1029 pKa = 5.14AGAITLKK1036 pKa = 11.0NNAGQTYY1043 pKa = 9.93NNSSSGNSDD1052 pKa = 2.9VVAAIEE1058 pKa = 4.21TDD1060 pKa = 2.99NGFQVLLEE1068 pKa = 4.3GTGSRR1073 pKa = 11.84NDD1075 pKa = 3.39TFYY1078 pKa = 11.64VWNTNSNGVITGGSGWKK1095 pKa = 10.22SGDD1098 pKa = 2.84IATRR1102 pKa = 11.84LGWEE1106 pKa = 3.9EE1107 pKa = 4.07TFNFDD1112 pKa = 3.69TNRR1115 pKa = 11.84DD1116 pKa = 3.52GTIGALILDD1125 pKa = 4.83DD1126 pKa = 5.09DD1127 pKa = 4.67NNGIVDD1133 pKa = 4.05GTEE1136 pKa = 3.5LTAYY1140 pKa = 10.44QLFSEE1145 pKa = 5.14AGAITLKK1152 pKa = 11.0NNAGQTYY1159 pKa = 9.93NNSSSGNSDD1168 pKa = 2.9VVAAIEE1174 pKa = 4.21TDD1176 pKa = 2.99NGFQVLLEE1184 pKa = 4.3GTGSRR1189 pKa = 11.84NDD1191 pKa = 3.39TFYY1194 pKa = 11.64VWNTNSNGVITGGSGWKK1211 pKa = 10.22SGDD1214 pKa = 2.84IATRR1218 pKa = 11.84LGWEE1222 pKa = 3.9EE1223 pKa = 4.07TFNFDD1228 pKa = 3.69TNRR1231 pKa = 11.84DD1232 pKa = 3.52GTIGALILDD1241 pKa = 4.83DD1242 pKa = 5.09DD1243 pKa = 4.67NNGIVDD1249 pKa = 4.05GTEE1252 pKa = 3.5LTAYY1256 pKa = 10.44QLFSEE1261 pKa = 5.14AGAITLKK1268 pKa = 11.0NNAGQTYY1275 pKa = 9.93NNSSSGNSDD1284 pKa = 2.9VVAAIEE1290 pKa = 4.21TDD1292 pKa = 2.99NGFQVLLEE1300 pKa = 4.3GTGSRR1305 pKa = 11.84NDD1307 pKa = 3.39TFYY1310 pKa = 11.62VWDD1313 pKa = 3.94TNSNGVITGGSGWKK1327 pKa = 9.67SGNIATRR1334 pKa = 11.84LGWEE1338 pKa = 3.9EE1339 pKa = 4.07TFNFDD1344 pKa = 3.69TNRR1347 pKa = 11.84DD1348 pKa = 3.61GTIGLPPSLSSPTLEE1363 pKa = 5.49ARR1365 pKa = 11.84FKK1367 pKa = 10.78DD1368 pKa = 3.47QWHH1371 pKa = 6.68LKK1373 pKa = 10.03DD1374 pKa = 4.03SSSGGANVASAWLLKK1389 pKa = 10.56NKK1391 pKa = 10.1SGSNIYY1397 pKa = 8.77GTGIHH1402 pKa = 6.59INVIDD1407 pKa = 5.64DD1408 pKa = 3.98GLDD1411 pKa = 3.03WRR1413 pKa = 11.84HH1414 pKa = 5.74QDD1416 pKa = 3.59LSTNYY1421 pKa = 9.32ISASSYY1427 pKa = 11.41DD1428 pKa = 3.95YY1429 pKa = 11.14VGKK1432 pKa = 10.56DD1433 pKa = 3.32NDD1435 pKa = 3.98PTPSSSADD1443 pKa = 3.0HH1444 pKa = 5.9GTAVAGVAAGYY1455 pKa = 8.67GHH1457 pKa = 6.88NGIGITGAAPNANISGQRR1475 pKa = 11.84LLGAGTARR1483 pKa = 11.84NEE1485 pKa = 3.67ASALTRR1491 pKa = 11.84TMNAVDD1497 pKa = 4.06IYY1499 pKa = 10.83SNSWGPNDD1507 pKa = 4.21NGRR1510 pKa = 11.84LQAAPARR1517 pKa = 11.84VLAALKK1523 pKa = 10.84DD1524 pKa = 3.79GVTNGRR1530 pKa = 11.84DD1531 pKa = 3.45GKK1533 pKa = 10.74GAIYY1537 pKa = 8.85TWAGGNGRR1545 pKa = 11.84NSNDD1549 pKa = 2.73NSNYY1553 pKa = 10.41DD1554 pKa = 3.06GYY1556 pKa = 11.73ANSRR1560 pKa = 11.84YY1561 pKa = 9.11VISVAAMTNRR1571 pKa = 11.84GRR1573 pKa = 11.84YY1574 pKa = 8.86SGYY1577 pKa = 9.99SEE1579 pKa = 4.74PGANVLVSAPSNGGTDD1595 pKa = 4.48AITTTSTNNSYY1606 pKa = 10.75IDD1608 pKa = 3.65NFGGTSSATPLVSGVIALMLEE1629 pKa = 4.42ANPNLTWRR1637 pKa = 11.84DD1638 pKa = 3.7VQHH1641 pKa = 6.33VLVNSSDD1648 pKa = 3.92VVDD1651 pKa = 4.38ASSNGWFTNGAEE1663 pKa = 4.44HH1664 pKa = 7.59DD1665 pKa = 4.13FSHH1668 pKa = 7.85DD1669 pKa = 3.57YY1670 pKa = 11.01GFGRR1674 pKa = 11.84INAEE1678 pKa = 3.74AAVALAKK1685 pKa = 9.29TWNNVGDD1692 pKa = 4.07EE1693 pKa = 4.29VSYY1696 pKa = 10.94SASTTPGIAIPDD1708 pKa = 3.67AGGGSISSTITISQDD1723 pKa = 2.37ITLEE1727 pKa = 4.22SVVIPILSDD1736 pKa = 2.87HH1737 pKa = 6.8TYY1739 pKa = 11.21AGDD1742 pKa = 3.55LTITLTSPEE1751 pKa = 4.2GTTAILSEE1759 pKa = 4.26GNRR1762 pKa = 11.84RR1763 pKa = 11.84DD1764 pKa = 3.44TSTLNFNFSAKK1775 pKa = 8.91TFWGEE1780 pKa = 3.65SSRR1783 pKa = 11.84GVWTLTINDD1792 pKa = 4.65LASLDD1797 pKa = 4.14TGTLDD1802 pKa = 3.18QWGLNLYY1809 pKa = 7.55GTQGTNFNAAITDD1822 pKa = 4.24SITGNEE1828 pKa = 4.26LEE1830 pKa = 4.65TFTGASGMTYY1840 pKa = 10.55ASKK1843 pKa = 10.56EE1844 pKa = 3.81ALEE1847 pKa = 4.19QIEE1850 pKa = 4.58GLSSYY1855 pKa = 10.46LSHH1858 pKa = 8.53IEE1860 pKa = 4.09DD1861 pKa = 4.66LLAEE1865 pKa = 4.46SPEE1868 pKa = 4.24KK1869 pKa = 10.44ATGDD1873 pKa = 3.66WLIGTQGPQDD1883 pKa = 3.51QIMRR1887 pKa = 11.84ASEE1890 pKa = 4.18LGISDD1895 pKa = 4.06SMTSQTFNAQNGISSFSMISALDD1918 pKa = 3.45PTEE1921 pKa = 5.83FIAQSLDD1928 pKa = 3.46SLGTNLSYY1936 pKa = 10.72AYY1938 pKa = 9.6PEE1940 pKa = 4.57LLHH1943 pKa = 5.99EE1944 pKa = 4.38TTTRR1948 pKa = 11.84SLIPSLEE1955 pKa = 3.97TSTFKK1960 pKa = 11.1AA1961 pKa = 3.7
MM1 pKa = 7.3NLSNLIEE8 pKa = 4.32SLSAYY13 pKa = 9.62RR14 pKa = 11.84ARR16 pKa = 11.84SLHH19 pKa = 5.77TSLNQPEE26 pKa = 4.82EE27 pKa = 4.37PDD29 pKa = 3.83PDD31 pKa = 3.47ISSIDD36 pKa = 3.73NNPALTPIDD45 pKa = 3.51QPLTVARR52 pKa = 11.84GLLQNGATTPPSQQLNSTEE71 pKa = 4.28SFSEE75 pKa = 4.52AINGNNSITTEE86 pKa = 4.03EE87 pKa = 4.3EE88 pKa = 3.87ANEE91 pKa = 4.0TTNIEE96 pKa = 3.8FRR98 pKa = 11.84IEE100 pKa = 4.09SKK102 pKa = 10.51YY103 pKa = 10.99LHH105 pKa = 7.01AGRR108 pKa = 11.84CACFACSYY116 pKa = 10.94SRR118 pKa = 11.84DD119 pKa = 3.56SLEE122 pKa = 4.11TNRR125 pKa = 11.84EE126 pKa = 4.25SNSEE130 pKa = 4.1LNNTLSNADD139 pKa = 3.38PTIAFGTLNEE149 pKa = 4.15LSDD152 pKa = 4.01YY153 pKa = 9.5LTTGFWEE160 pKa = 4.19EE161 pKa = 3.61AGTYY165 pKa = 7.19TRR167 pKa = 11.84RR168 pKa = 11.84FNLGGSGLGAKK179 pKa = 9.84NGQLTYY185 pKa = 10.99NITGWADD192 pKa = 3.34DD193 pKa = 4.12SNGLSAEE200 pKa = 4.16RR201 pKa = 11.84QNLTRR206 pKa = 11.84EE207 pKa = 4.05VFNVYY212 pKa = 9.39EE213 pKa = 5.0AITGIEE219 pKa = 4.12FVEE222 pKa = 4.71VATEE226 pKa = 3.88GDD228 pKa = 4.31FRR230 pKa = 11.84FTDD233 pKa = 3.74NDD235 pKa = 3.64SGAYY239 pKa = 9.83AYY241 pKa = 10.81LGGGWYY247 pKa = 9.94DD248 pKa = 3.29QDD250 pKa = 4.71PNTGTIDD257 pKa = 3.46YY258 pKa = 9.12NAAIIDD264 pKa = 3.69YY265 pKa = 10.57SVINVASSWFYY276 pKa = 11.49GDD278 pKa = 3.68SSYY281 pKa = 11.9NSYY284 pKa = 9.32TPQTIFHH291 pKa = 7.24EE292 pKa = 4.72IGHH295 pKa = 6.51ALGLGHH301 pKa = 6.58QGQYY305 pKa = 10.39NAGNGNPTYY314 pKa = 10.38EE315 pKa = 4.43NSAQYY320 pKa = 11.53GNDD323 pKa = 2.93TWLTTMMSYY332 pKa = 9.87WSQTTNTNVNASRR345 pKa = 11.84AYY347 pKa = 10.07LQTPMTVDD355 pKa = 3.9WIALDD360 pKa = 3.81KK361 pKa = 11.2LYY363 pKa = 11.05GSQGYY368 pKa = 10.19GSFKK372 pKa = 10.64AFNGNTVYY380 pKa = 10.78GVGTNISSGTSEE392 pKa = 3.97IMNNFATMISDD403 pKa = 3.9TAYY406 pKa = 9.87TLVDD410 pKa = 3.66GSGYY414 pKa = 10.79DD415 pKa = 3.2ILNVSNYY422 pKa = 10.35SDD424 pKa = 3.48DD425 pKa = 3.68QFINLAPSEE434 pKa = 4.31LNGTLPSSSSIGGLVNNLTIGVGTILEE461 pKa = 4.17EE462 pKa = 4.95AIGGSGNDD470 pKa = 3.29TFLGNIADD478 pKa = 3.8NVFRR482 pKa = 11.84GGNGGDD488 pKa = 3.07RR489 pKa = 11.84FYY491 pKa = 11.54DD492 pKa = 3.5SFGSDD497 pKa = 2.67TYY499 pKa = 11.41YY500 pKa = 11.06GGSGTFDD507 pKa = 2.76TLYY510 pKa = 10.81FSEE513 pKa = 4.75NYY515 pKa = 10.3SDD517 pKa = 5.56FSIEE521 pKa = 4.03NLGTSLAFSRR531 pKa = 11.84NNSGLADD538 pKa = 4.04TDD540 pKa = 5.5LIWNDD545 pKa = 2.73IEE547 pKa = 5.2LVYY550 pKa = 11.06FNDD553 pKa = 4.72DD554 pKa = 3.55VIKK557 pKa = 9.85TYY559 pKa = 10.83QEE561 pKa = 3.9LLADD565 pKa = 3.95ITPINTPPTANDD577 pKa = 3.1VFVATSEE584 pKa = 4.37DD585 pKa = 4.05SNSVSGNFNANDD597 pKa = 4.43PDD599 pKa = 4.47SSDD602 pKa = 3.1QLTYY606 pKa = 10.73TITTAPTIGSVVVNSDD622 pKa = 3.12GTFTYY627 pKa = 11.06NFGNNFQSLGDD638 pKa = 3.87GDD640 pKa = 3.92STAVVFQYY648 pKa = 10.52IASDD652 pKa = 3.99DD653 pKa = 4.09NATDD657 pKa = 3.68SAPATVTITINGSNDD672 pKa = 2.91APILSGTPSALTNGTEE688 pKa = 4.67DD689 pKa = 3.05IAYY692 pKa = 10.01LLKK695 pKa = 10.96ASDD698 pKa = 4.24LLQGYY703 pKa = 9.81SDD705 pKa = 3.89IDD707 pKa = 3.94GDD709 pKa = 4.18TLSITSVTTASANGTLTPNGNGTWTYY735 pKa = 11.08TPTQDD740 pKa = 4.41LNGEE744 pKa = 4.46VVFSFVVSDD753 pKa = 4.19GNGGTANGSTSLTLDD768 pKa = 3.49PVEE771 pKa = 5.89DD772 pKa = 3.84EE773 pKa = 4.64VLLDD777 pKa = 3.78EE778 pKa = 5.44DD779 pKa = 4.05NNGIVDD785 pKa = 4.06GTEE788 pKa = 3.5LTAYY792 pKa = 10.44QLFSEE797 pKa = 5.14AGAITLKK804 pKa = 11.0NNAGQTYY811 pKa = 9.93NNSSSGNSDD820 pKa = 2.9VVAAIEE826 pKa = 4.21TDD828 pKa = 2.99NGFQVLLEE836 pKa = 4.3GTGSRR841 pKa = 11.84NDD843 pKa = 3.39TFYY846 pKa = 11.64VWNTNSNGVIIGGSGWKK863 pKa = 10.22SGDD866 pKa = 2.84IATRR870 pKa = 11.84LGWEE874 pKa = 3.9EE875 pKa = 4.07TFNFDD880 pKa = 3.69TNRR883 pKa = 11.84DD884 pKa = 3.52GTIGALILDD893 pKa = 4.83DD894 pKa = 5.09DD895 pKa = 4.67NNGIVDD901 pKa = 4.05GTEE904 pKa = 3.5LTAYY908 pKa = 10.44QLFSEE913 pKa = 5.14AGAITLKK920 pKa = 11.0NNAGQTYY927 pKa = 9.93NNSSSGNSDD936 pKa = 2.9VVAAIEE942 pKa = 4.21TDD944 pKa = 2.99NGFQVLLEE952 pKa = 4.3GTGSRR957 pKa = 11.84NDD959 pKa = 3.39TFYY962 pKa = 11.64VWNTNSNGVIIGGSGWKK979 pKa = 10.22SGDD982 pKa = 2.84IATRR986 pKa = 11.84LGWEE990 pKa = 3.9EE991 pKa = 4.07TFNFDD996 pKa = 3.69TNRR999 pKa = 11.84DD1000 pKa = 3.52GTIGALILDD1009 pKa = 4.83DD1010 pKa = 5.09DD1011 pKa = 4.67NNGIVDD1017 pKa = 4.05GTEE1020 pKa = 3.5LTAYY1024 pKa = 10.44QLFSEE1029 pKa = 5.14AGAITLKK1036 pKa = 11.0NNAGQTYY1043 pKa = 9.93NNSSSGNSDD1052 pKa = 2.9VVAAIEE1058 pKa = 4.21TDD1060 pKa = 2.99NGFQVLLEE1068 pKa = 4.3GTGSRR1073 pKa = 11.84NDD1075 pKa = 3.39TFYY1078 pKa = 11.64VWNTNSNGVITGGSGWKK1095 pKa = 10.22SGDD1098 pKa = 2.84IATRR1102 pKa = 11.84LGWEE1106 pKa = 3.9EE1107 pKa = 4.07TFNFDD1112 pKa = 3.69TNRR1115 pKa = 11.84DD1116 pKa = 3.52GTIGALILDD1125 pKa = 4.83DD1126 pKa = 5.09DD1127 pKa = 4.67NNGIVDD1133 pKa = 4.05GTEE1136 pKa = 3.5LTAYY1140 pKa = 10.44QLFSEE1145 pKa = 5.14AGAITLKK1152 pKa = 11.0NNAGQTYY1159 pKa = 9.93NNSSSGNSDD1168 pKa = 2.9VVAAIEE1174 pKa = 4.21TDD1176 pKa = 2.99NGFQVLLEE1184 pKa = 4.3GTGSRR1189 pKa = 11.84NDD1191 pKa = 3.39TFYY1194 pKa = 11.64VWNTNSNGVITGGSGWKK1211 pKa = 10.22SGDD1214 pKa = 2.84IATRR1218 pKa = 11.84LGWEE1222 pKa = 3.9EE1223 pKa = 4.07TFNFDD1228 pKa = 3.69TNRR1231 pKa = 11.84DD1232 pKa = 3.52GTIGALILDD1241 pKa = 4.83DD1242 pKa = 5.09DD1243 pKa = 4.67NNGIVDD1249 pKa = 4.05GTEE1252 pKa = 3.5LTAYY1256 pKa = 10.44QLFSEE1261 pKa = 5.14AGAITLKK1268 pKa = 11.0NNAGQTYY1275 pKa = 9.93NNSSSGNSDD1284 pKa = 2.9VVAAIEE1290 pKa = 4.21TDD1292 pKa = 2.99NGFQVLLEE1300 pKa = 4.3GTGSRR1305 pKa = 11.84NDD1307 pKa = 3.39TFYY1310 pKa = 11.62VWDD1313 pKa = 3.94TNSNGVITGGSGWKK1327 pKa = 9.67SGNIATRR1334 pKa = 11.84LGWEE1338 pKa = 3.9EE1339 pKa = 4.07TFNFDD1344 pKa = 3.69TNRR1347 pKa = 11.84DD1348 pKa = 3.61GTIGLPPSLSSPTLEE1363 pKa = 5.49ARR1365 pKa = 11.84FKK1367 pKa = 10.78DD1368 pKa = 3.47QWHH1371 pKa = 6.68LKK1373 pKa = 10.03DD1374 pKa = 4.03SSSGGANVASAWLLKK1389 pKa = 10.56NKK1391 pKa = 10.1SGSNIYY1397 pKa = 8.77GTGIHH1402 pKa = 6.59INVIDD1407 pKa = 5.64DD1408 pKa = 3.98GLDD1411 pKa = 3.03WRR1413 pKa = 11.84HH1414 pKa = 5.74QDD1416 pKa = 3.59LSTNYY1421 pKa = 9.32ISASSYY1427 pKa = 11.41DD1428 pKa = 3.95YY1429 pKa = 11.14VGKK1432 pKa = 10.56DD1433 pKa = 3.32NDD1435 pKa = 3.98PTPSSSADD1443 pKa = 3.0HH1444 pKa = 5.9GTAVAGVAAGYY1455 pKa = 8.67GHH1457 pKa = 6.88NGIGITGAAPNANISGQRR1475 pKa = 11.84LLGAGTARR1483 pKa = 11.84NEE1485 pKa = 3.67ASALTRR1491 pKa = 11.84TMNAVDD1497 pKa = 4.06IYY1499 pKa = 10.83SNSWGPNDD1507 pKa = 4.21NGRR1510 pKa = 11.84LQAAPARR1517 pKa = 11.84VLAALKK1523 pKa = 10.84DD1524 pKa = 3.79GVTNGRR1530 pKa = 11.84DD1531 pKa = 3.45GKK1533 pKa = 10.74GAIYY1537 pKa = 8.85TWAGGNGRR1545 pKa = 11.84NSNDD1549 pKa = 2.73NSNYY1553 pKa = 10.41DD1554 pKa = 3.06GYY1556 pKa = 11.73ANSRR1560 pKa = 11.84YY1561 pKa = 9.11VISVAAMTNRR1571 pKa = 11.84GRR1573 pKa = 11.84YY1574 pKa = 8.86SGYY1577 pKa = 9.99SEE1579 pKa = 4.74PGANVLVSAPSNGGTDD1595 pKa = 4.48AITTTSTNNSYY1606 pKa = 10.75IDD1608 pKa = 3.65NFGGTSSATPLVSGVIALMLEE1629 pKa = 4.42ANPNLTWRR1637 pKa = 11.84DD1638 pKa = 3.7VQHH1641 pKa = 6.33VLVNSSDD1648 pKa = 3.92VVDD1651 pKa = 4.38ASSNGWFTNGAEE1663 pKa = 4.44HH1664 pKa = 7.59DD1665 pKa = 4.13FSHH1668 pKa = 7.85DD1669 pKa = 3.57YY1670 pKa = 11.01GFGRR1674 pKa = 11.84INAEE1678 pKa = 3.74AAVALAKK1685 pKa = 9.29TWNNVGDD1692 pKa = 4.07EE1693 pKa = 4.29VSYY1696 pKa = 10.94SASTTPGIAIPDD1708 pKa = 3.67AGGGSISSTITISQDD1723 pKa = 2.37ITLEE1727 pKa = 4.22SVVIPILSDD1736 pKa = 2.87HH1737 pKa = 6.8TYY1739 pKa = 11.21AGDD1742 pKa = 3.55LTITLTSPEE1751 pKa = 4.2GTTAILSEE1759 pKa = 4.26GNRR1762 pKa = 11.84RR1763 pKa = 11.84DD1764 pKa = 3.44TSTLNFNFSAKK1775 pKa = 8.91TFWGEE1780 pKa = 3.65SSRR1783 pKa = 11.84GVWTLTINDD1792 pKa = 4.65LASLDD1797 pKa = 4.14TGTLDD1802 pKa = 3.18QWGLNLYY1809 pKa = 7.55GTQGTNFNAAITDD1822 pKa = 4.24SITGNEE1828 pKa = 4.26LEE1830 pKa = 4.65TFTGASGMTYY1840 pKa = 10.55ASKK1843 pKa = 10.56EE1844 pKa = 3.81ALEE1847 pKa = 4.19QIEE1850 pKa = 4.58GLSSYY1855 pKa = 10.46LSHH1858 pKa = 8.53IEE1860 pKa = 4.09DD1861 pKa = 4.66LLAEE1865 pKa = 4.46SPEE1868 pKa = 4.24KK1869 pKa = 10.44ATGDD1873 pKa = 3.66WLIGTQGPQDD1883 pKa = 3.51QIMRR1887 pKa = 11.84ASEE1890 pKa = 4.18LGISDD1895 pKa = 4.06SMTSQTFNAQNGISSFSMISALDD1918 pKa = 3.45PTEE1921 pKa = 5.83FIAQSLDD1928 pKa = 3.46SLGTNLSYY1936 pKa = 10.72AYY1938 pKa = 9.6PEE1940 pKa = 4.57LLHH1943 pKa = 5.99EE1944 pKa = 4.38TTTRR1948 pKa = 11.84SLIPSLEE1955 pKa = 3.97TSTFKK1960 pKa = 11.1AA1961 pKa = 3.7
Molecular weight: 208.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4CRW8|A4CRW8_SYNPV Uncharacterized protein OS=Synechococcus sp. (strain WH7805) OX=59931 GN=WH7805_13833 PE=4 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.07RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 6.21TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84TRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.66RR38 pKa = 11.84GRR40 pKa = 11.84SRR42 pKa = 11.84LSVV45 pKa = 3.08
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.07RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 6.21TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84TRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.66RR38 pKa = 11.84GRR40 pKa = 11.84SRR42 pKa = 11.84LSVV45 pKa = 3.08
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
787820 |
20 |
8129 |
274.1 |
29.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.107 ± 0.064 | 1.226 ± 0.027 |
5.66 ± 0.074 | 5.659 ± 0.063 |
3.2 ± 0.042 | 8.267 ± 0.06 |
2.07 ± 0.045 | 4.456 ± 0.05 |
2.73 ± 0.044 | 12.315 ± 0.098 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.066 ± 0.033 | 2.945 ± 0.086 |
5.367 ± 0.063 | 4.614 ± 0.056 |
7.016 ± 0.098 | 6.65 ± 0.067 |
5.133 ± 0.147 | 6.998 ± 0.046 |
1.721 ± 0.035 | 1.801 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
