
Dictyobacter aurantiacus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Ktedonobacteria; Ktedonobacterales; Dictyobacteraceae; Dictyobacter
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
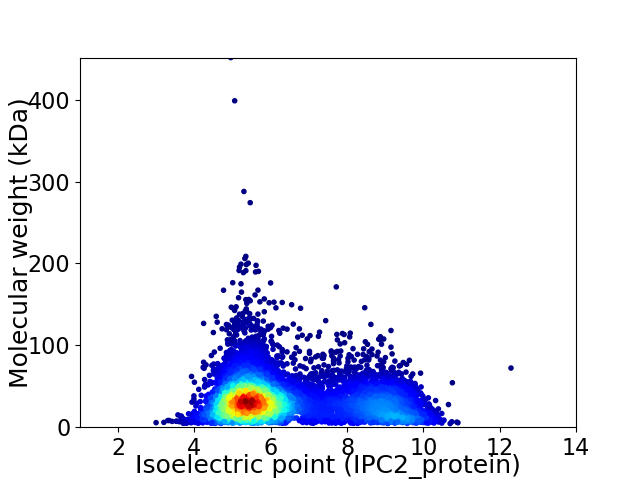
Virtual 2D-PAGE plot for 7362 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
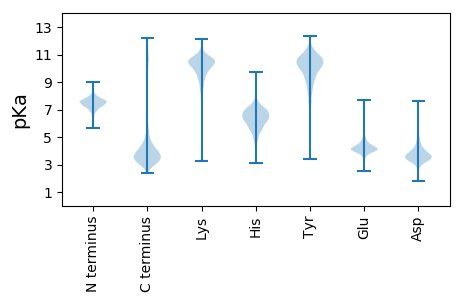
Protein with the lowest isoelectric point:
>tr|A0A401ZJZ6|A0A401ZJZ6_9CHLR Histidine kinase OS=Dictyobacter aurantiacus OX=1936993 GN=KDAU_45050 PE=4 SV=1
MM1 pKa = 7.47SVTPSPTVVGVFRR14 pKa = 11.84DD15 pKa = 3.58RR16 pKa = 11.84SVVEE20 pKa = 3.74QVIEE24 pKa = 3.96ALYY27 pKa = 10.34QAGFAQDD34 pKa = 3.43QIRR37 pKa = 11.84YY38 pKa = 8.4SVPDD42 pKa = 3.45NTSGGFLNDD51 pKa = 3.7LKK53 pKa = 11.28HH54 pKa = 6.85LFTGPNASGSGDD66 pKa = 3.69LANDD70 pKa = 3.6LTGMGLTSEE79 pKa = 4.21QARR82 pKa = 11.84YY83 pKa = 8.5YY84 pKa = 10.86ANEE87 pKa = 3.92YY88 pKa = 11.07DD89 pKa = 3.49NGNIVMAVQALDD101 pKa = 3.88NEE103 pKa = 4.34QQAMNIFHH111 pKa = 6.51QFGAYY116 pKa = 9.98GIHH119 pKa = 5.22NTLRR123 pKa = 11.84EE124 pKa = 4.26AEE126 pKa = 4.38GMNEE130 pKa = 4.02DD131 pKa = 4.0AGYY134 pKa = 10.1AQPQTDD140 pKa = 4.15FNNPSAAYY148 pKa = 8.73AQQSTDD154 pKa = 3.46YY155 pKa = 8.91QHH157 pKa = 6.62QMSPYY162 pKa = 7.55TQHH165 pKa = 7.11PEE167 pKa = 4.64DD168 pKa = 4.63YY169 pKa = 9.34AQQPASSTWGNNAEE183 pKa = 4.51HH184 pKa = 6.24GQPLNEE190 pKa = 4.17EE191 pKa = 4.11TNPRR195 pKa = 11.84QEE197 pKa = 5.05AVTNDD202 pKa = 3.07QTAVHH207 pKa = 6.99EE208 pKa = 4.23YY209 pKa = 11.32ANDD212 pKa = 3.34NNYY215 pKa = 9.64TYY217 pKa = 11.27NDD219 pKa = 3.1NAAYY223 pKa = 10.98NNDD226 pKa = 2.8NTAYY230 pKa = 9.97NDD232 pKa = 3.46SNAEE236 pKa = 4.05YY237 pKa = 10.82NEE239 pKa = 3.81NNADD243 pKa = 3.63YY244 pKa = 10.17TEE246 pKa = 4.27QNSTVAPAGAADD258 pKa = 4.87QRR260 pKa = 11.84DD261 pKa = 4.31LPDD264 pKa = 4.28NIQPEE269 pKa = 4.77SVVDD273 pKa = 3.66HH274 pKa = 5.51QQNLPEE280 pKa = 4.51DD281 pKa = 3.79TTPIRR286 pKa = 11.84EE287 pKa = 3.93PSVSDD292 pKa = 3.25EE293 pKa = 4.35HH294 pKa = 8.38LNAPDD299 pKa = 4.28HH300 pKa = 6.42QDD302 pKa = 3.37QAAQDD307 pKa = 3.74YY308 pKa = 9.61RR309 pKa = 11.84SPEE312 pKa = 4.12TEE314 pKa = 4.73DD315 pKa = 5.3DD316 pKa = 3.75PTADD320 pKa = 3.62RR321 pKa = 11.84QDD323 pKa = 3.73YY324 pKa = 11.19NSADD328 pKa = 3.67NQGDD332 pKa = 3.49ITDD335 pKa = 3.93EE336 pKa = 4.14TTQPVGSDD344 pKa = 3.38DD345 pKa = 3.37RR346 pKa = 11.84AGAYY350 pKa = 7.78RR351 pKa = 11.84APATDD356 pKa = 4.26SDD358 pKa = 4.23TQNATSAPVIDD369 pKa = 4.45GANRR373 pKa = 11.84EE374 pKa = 4.22YY375 pKa = 9.41PTTVTYY381 pKa = 10.88DD382 pKa = 3.56EE383 pKa = 4.86TPGTPAADD391 pKa = 3.85NAPLDD396 pKa = 4.15AQPANAVDD404 pKa = 3.45NSTDD408 pKa = 3.32NTTSTAQHH416 pKa = 6.41TDD418 pKa = 2.82TFEE421 pKa = 3.9NAAYY425 pKa = 9.92QSPATHH431 pKa = 6.34NGEE434 pKa = 4.15VATLSTNYY442 pKa = 8.43NTEE445 pKa = 4.09PDD447 pKa = 3.51LSADD451 pKa = 3.18QGQIDD456 pKa = 4.22EE457 pKa = 5.47ADD459 pKa = 3.34HH460 pKa = 5.86QQYY463 pKa = 7.53QTAASTTEE471 pKa = 3.95NAPLDD476 pKa = 3.96QQQAPQDD483 pKa = 3.69ATAGTTATDD492 pKa = 4.33FAAQRR497 pKa = 11.84EE498 pKa = 4.27EE499 pKa = 4.44RR500 pKa = 11.84LRR502 pKa = 11.84QLQQRR507 pKa = 11.84LEE509 pKa = 4.16SAKK512 pKa = 10.2QRR514 pKa = 11.84LQDD517 pKa = 4.09SQTQLQAAKK526 pKa = 9.22EE527 pKa = 4.25HH528 pKa = 6.36EE529 pKa = 4.47SQLQSMKK536 pKa = 10.36QQLDD540 pKa = 3.79AIEE543 pKa = 5.32AEE545 pKa = 4.28LEE547 pKa = 4.13KK548 pKa = 11.41NEE550 pKa = 3.96AEE552 pKa = 4.55LRR554 pKa = 11.84EE555 pKa = 3.92THH557 pKa = 6.91ARR559 pKa = 11.84IEE561 pKa = 3.89QYY563 pKa = 10.78YY564 pKa = 10.14
MM1 pKa = 7.47SVTPSPTVVGVFRR14 pKa = 11.84DD15 pKa = 3.58RR16 pKa = 11.84SVVEE20 pKa = 3.74QVIEE24 pKa = 3.96ALYY27 pKa = 10.34QAGFAQDD34 pKa = 3.43QIRR37 pKa = 11.84YY38 pKa = 8.4SVPDD42 pKa = 3.45NTSGGFLNDD51 pKa = 3.7LKK53 pKa = 11.28HH54 pKa = 6.85LFTGPNASGSGDD66 pKa = 3.69LANDD70 pKa = 3.6LTGMGLTSEE79 pKa = 4.21QARR82 pKa = 11.84YY83 pKa = 8.5YY84 pKa = 10.86ANEE87 pKa = 3.92YY88 pKa = 11.07DD89 pKa = 3.49NGNIVMAVQALDD101 pKa = 3.88NEE103 pKa = 4.34QQAMNIFHH111 pKa = 6.51QFGAYY116 pKa = 9.98GIHH119 pKa = 5.22NTLRR123 pKa = 11.84EE124 pKa = 4.26AEE126 pKa = 4.38GMNEE130 pKa = 4.02DD131 pKa = 4.0AGYY134 pKa = 10.1AQPQTDD140 pKa = 4.15FNNPSAAYY148 pKa = 8.73AQQSTDD154 pKa = 3.46YY155 pKa = 8.91QHH157 pKa = 6.62QMSPYY162 pKa = 7.55TQHH165 pKa = 7.11PEE167 pKa = 4.64DD168 pKa = 4.63YY169 pKa = 9.34AQQPASSTWGNNAEE183 pKa = 4.51HH184 pKa = 6.24GQPLNEE190 pKa = 4.17EE191 pKa = 4.11TNPRR195 pKa = 11.84QEE197 pKa = 5.05AVTNDD202 pKa = 3.07QTAVHH207 pKa = 6.99EE208 pKa = 4.23YY209 pKa = 11.32ANDD212 pKa = 3.34NNYY215 pKa = 9.64TYY217 pKa = 11.27NDD219 pKa = 3.1NAAYY223 pKa = 10.98NNDD226 pKa = 2.8NTAYY230 pKa = 9.97NDD232 pKa = 3.46SNAEE236 pKa = 4.05YY237 pKa = 10.82NEE239 pKa = 3.81NNADD243 pKa = 3.63YY244 pKa = 10.17TEE246 pKa = 4.27QNSTVAPAGAADD258 pKa = 4.87QRR260 pKa = 11.84DD261 pKa = 4.31LPDD264 pKa = 4.28NIQPEE269 pKa = 4.77SVVDD273 pKa = 3.66HH274 pKa = 5.51QQNLPEE280 pKa = 4.51DD281 pKa = 3.79TTPIRR286 pKa = 11.84EE287 pKa = 3.93PSVSDD292 pKa = 3.25EE293 pKa = 4.35HH294 pKa = 8.38LNAPDD299 pKa = 4.28HH300 pKa = 6.42QDD302 pKa = 3.37QAAQDD307 pKa = 3.74YY308 pKa = 9.61RR309 pKa = 11.84SPEE312 pKa = 4.12TEE314 pKa = 4.73DD315 pKa = 5.3DD316 pKa = 3.75PTADD320 pKa = 3.62RR321 pKa = 11.84QDD323 pKa = 3.73YY324 pKa = 11.19NSADD328 pKa = 3.67NQGDD332 pKa = 3.49ITDD335 pKa = 3.93EE336 pKa = 4.14TTQPVGSDD344 pKa = 3.38DD345 pKa = 3.37RR346 pKa = 11.84AGAYY350 pKa = 7.78RR351 pKa = 11.84APATDD356 pKa = 4.26SDD358 pKa = 4.23TQNATSAPVIDD369 pKa = 4.45GANRR373 pKa = 11.84EE374 pKa = 4.22YY375 pKa = 9.41PTTVTYY381 pKa = 10.88DD382 pKa = 3.56EE383 pKa = 4.86TPGTPAADD391 pKa = 3.85NAPLDD396 pKa = 4.15AQPANAVDD404 pKa = 3.45NSTDD408 pKa = 3.32NTTSTAQHH416 pKa = 6.41TDD418 pKa = 2.82TFEE421 pKa = 3.9NAAYY425 pKa = 9.92QSPATHH431 pKa = 6.34NGEE434 pKa = 4.15VATLSTNYY442 pKa = 8.43NTEE445 pKa = 4.09PDD447 pKa = 3.51LSADD451 pKa = 3.18QGQIDD456 pKa = 4.22EE457 pKa = 5.47ADD459 pKa = 3.34HH460 pKa = 5.86QQYY463 pKa = 7.53QTAASTTEE471 pKa = 3.95NAPLDD476 pKa = 3.96QQQAPQDD483 pKa = 3.69ATAGTTATDD492 pKa = 4.33FAAQRR497 pKa = 11.84EE498 pKa = 4.27EE499 pKa = 4.44RR500 pKa = 11.84LRR502 pKa = 11.84QLQQRR507 pKa = 11.84LEE509 pKa = 4.16SAKK512 pKa = 10.2QRR514 pKa = 11.84LQDD517 pKa = 4.09SQTQLQAAKK526 pKa = 9.22EE527 pKa = 4.25HH528 pKa = 6.36EE529 pKa = 4.47SQLQSMKK536 pKa = 10.36QQLDD540 pKa = 3.79AIEE543 pKa = 5.32AEE545 pKa = 4.28LEE547 pKa = 4.13KK548 pKa = 11.41NEE550 pKa = 3.96AEE552 pKa = 4.55LRR554 pKa = 11.84EE555 pKa = 3.92THH557 pKa = 6.91ARR559 pKa = 11.84IEE561 pKa = 3.89QYY563 pKa = 10.78YY564 pKa = 10.14
Molecular weight: 61.99 kDa
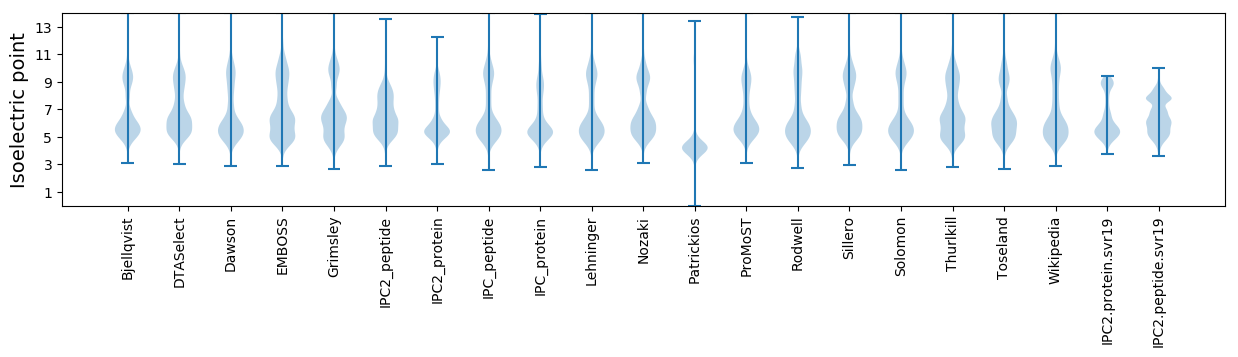
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A401ZEE7|A0A401ZEE7_9CHLR Enterochelin esterase OS=Dictyobacter aurantiacus OX=1936993 GN=fes PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.45RR3 pKa = 11.84TWQPKK8 pKa = 8.26RR9 pKa = 11.84IPRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84EE15 pKa = 3.41HH16 pKa = 7.15GFMKK20 pKa = 10.47RR21 pKa = 11.84MSTRR25 pKa = 11.84NGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.49ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.41GRR39 pKa = 11.84WSLTVV44 pKa = 3.02
MM1 pKa = 7.35KK2 pKa = 9.45RR3 pKa = 11.84TWQPKK8 pKa = 8.26RR9 pKa = 11.84IPRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84EE15 pKa = 3.41HH16 pKa = 7.15GFMKK20 pKa = 10.47RR21 pKa = 11.84MSTRR25 pKa = 11.84NGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.49ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.41GRR39 pKa = 11.84WSLTVV44 pKa = 3.02
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2368895 |
39 |
4032 |
321.8 |
35.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.439 ± 0.029 | 0.978 ± 0.009 |
4.904 ± 0.022 | 5.77 ± 0.036 |
3.612 ± 0.02 | 7.149 ± 0.026 |
2.619 ± 0.014 | 5.853 ± 0.024 |
3.198 ± 0.021 | 10.847 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.349 ± 0.012 | 3.323 ± 0.022 |
5.168 ± 0.024 | 5.167 ± 0.023 |
6.238 ± 0.028 | 6.129 ± 0.026 |
5.858 ± 0.023 | 6.77 ± 0.027 |
1.494 ± 0.013 | 3.134 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
