
Strawberry mild yellow edge-associated virus (SMYEaV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Potexvirus
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
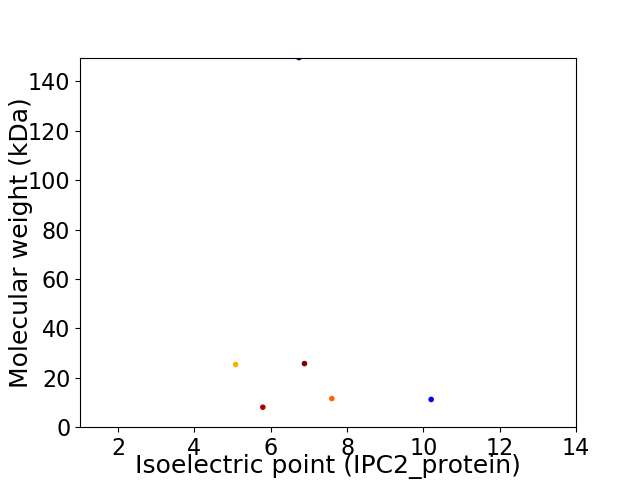
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
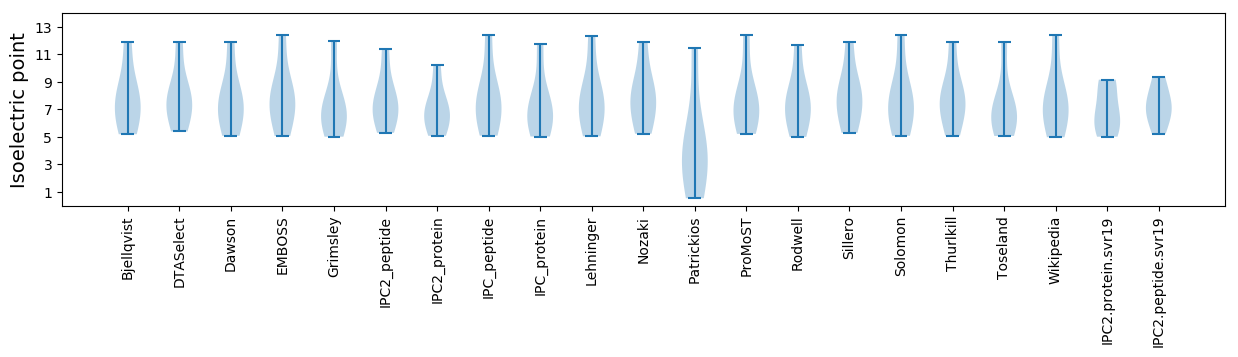
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q00848|TGB3_SMYEA Movement protein TGBp3 OS=Strawberry mild yellow edge-associated virus OX=12187 GN=ORF4 PE=3 SV=1
MM1 pKa = 8.47VEE3 pKa = 3.87FTKK6 pKa = 10.79RR7 pKa = 11.84LLLEE11 pKa = 4.44RR12 pKa = 11.84NFEE15 pKa = 4.1RR16 pKa = 11.84TNRR19 pKa = 11.84PITGPIVVHH28 pKa = 6.33AVAGAGKK35 pKa = 10.02SSVINTVSLTFQLICWTTLPEE56 pKa = 4.06EE57 pKa = 4.33KK58 pKa = 10.64ASFNCLHH65 pKa = 6.46LRR67 pKa = 11.84HH68 pKa = 6.62LDD70 pKa = 3.59GPAFPGAFVDD80 pKa = 5.08EE81 pKa = 4.46YY82 pKa = 11.61QLADD86 pKa = 3.54TDD88 pKa = 4.0LSEE91 pKa = 4.38AAFLFGDD98 pKa = 4.15PLQYY102 pKa = 10.1PGPAAQVPHH111 pKa = 6.03FVKK114 pKa = 10.61LFSHH118 pKa = 6.69RR119 pKa = 11.84CGLNSASLIRR129 pKa = 11.84EE130 pKa = 3.9LGIAFEE136 pKa = 4.12ASKK139 pKa = 10.81LDD141 pKa = 3.92SVQHH145 pKa = 6.45LDD147 pKa = 4.18PYY149 pKa = 11.25SSDD152 pKa = 3.6PEE154 pKa = 4.16GTILAFEE161 pKa = 4.95PEE163 pKa = 4.32VQAALASHH171 pKa = 6.61SLDD174 pKa = 3.66FLCLDD179 pKa = 4.26EE180 pKa = 5.84FRR182 pKa = 11.84GKK184 pKa = 9.04QWPVCTLYY192 pKa = 11.21VSTKK196 pKa = 10.11NLCDD200 pKa = 3.92LDD202 pKa = 4.25RR203 pKa = 11.84PSVYY207 pKa = 10.35VALTRR212 pKa = 11.84HH213 pKa = 5.36YY214 pKa = 10.41EE215 pKa = 3.84RR216 pKa = 11.84LLIMSFDD223 pKa = 3.99AADD226 pKa = 3.57TSAA229 pKa = 4.49
MM1 pKa = 8.47VEE3 pKa = 3.87FTKK6 pKa = 10.79RR7 pKa = 11.84LLLEE11 pKa = 4.44RR12 pKa = 11.84NFEE15 pKa = 4.1RR16 pKa = 11.84TNRR19 pKa = 11.84PITGPIVVHH28 pKa = 6.33AVAGAGKK35 pKa = 10.02SSVINTVSLTFQLICWTTLPEE56 pKa = 4.06EE57 pKa = 4.33KK58 pKa = 10.64ASFNCLHH65 pKa = 6.46LRR67 pKa = 11.84HH68 pKa = 6.62LDD70 pKa = 3.59GPAFPGAFVDD80 pKa = 5.08EE81 pKa = 4.46YY82 pKa = 11.61QLADD86 pKa = 3.54TDD88 pKa = 4.0LSEE91 pKa = 4.38AAFLFGDD98 pKa = 4.15PLQYY102 pKa = 10.1PGPAAQVPHH111 pKa = 6.03FVKK114 pKa = 10.61LFSHH118 pKa = 6.69RR119 pKa = 11.84CGLNSASLIRR129 pKa = 11.84EE130 pKa = 3.9LGIAFEE136 pKa = 4.12ASKK139 pKa = 10.81LDD141 pKa = 3.92SVQHH145 pKa = 6.45LDD147 pKa = 4.18PYY149 pKa = 11.25SSDD152 pKa = 3.6PEE154 pKa = 4.16GTILAFEE161 pKa = 4.95PEE163 pKa = 4.32VQAALASHH171 pKa = 6.61SLDD174 pKa = 3.66FLCLDD179 pKa = 4.26EE180 pKa = 5.84FRR182 pKa = 11.84GKK184 pKa = 9.04QWPVCTLYY192 pKa = 11.21VSTKK196 pKa = 10.11NLCDD200 pKa = 3.92LDD202 pKa = 4.25RR203 pKa = 11.84PSVYY207 pKa = 10.35VALTRR212 pKa = 11.84HH213 pKa = 5.36YY214 pKa = 10.41EE215 pKa = 3.84RR216 pKa = 11.84LLIMSFDD223 pKa = 3.99AADD226 pKa = 3.57TSAA229 pKa = 4.49
Molecular weight: 25.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q88468|Q88468_SMYEA 11K protein OS=Strawberry mild yellow edge-associated virus OX=12187 PE=4 SV=1
MM1 pKa = 7.29TLALHH6 pKa = 6.68PLPNRR11 pKa = 11.84SVLHH15 pKa = 6.12PSAARR20 pKa = 11.84ACKK23 pKa = 10.16SPPSFEE29 pKa = 3.81ITALSASSVCSTPQASGTKK48 pKa = 9.41QSGTTVPLAIGAISSLHH65 pKa = 6.02RR66 pKa = 11.84RR67 pKa = 11.84PSSLPLISSMVCSIQPAKK85 pKa = 10.37KK86 pKa = 10.02SRR88 pKa = 11.84FGVSPPPRR96 pKa = 11.84RR97 pKa = 11.84FMPQPRR103 pKa = 11.84IRR105 pKa = 11.84MM106 pKa = 3.79
MM1 pKa = 7.29TLALHH6 pKa = 6.68PLPNRR11 pKa = 11.84SVLHH15 pKa = 6.12PSAARR20 pKa = 11.84ACKK23 pKa = 10.16SPPSFEE29 pKa = 3.81ITALSASSVCSTPQASGTKK48 pKa = 9.41QSGTTVPLAIGAISSLHH65 pKa = 6.02RR66 pKa = 11.84RR67 pKa = 11.84PSSLPLISSMVCSIQPAKK85 pKa = 10.37KK86 pKa = 10.02SRR88 pKa = 11.84FGVSPPPRR96 pKa = 11.84RR97 pKa = 11.84FMPQPRR103 pKa = 11.84IRR105 pKa = 11.84MM106 pKa = 3.79
Molecular weight: 11.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2083 |
75 |
1323 |
347.2 |
38.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.785 ± 0.787 | 2.064 ± 0.496 |
5.713 ± 0.856 | 4.897 ± 1.034 |
4.273 ± 0.363 | 5.233 ± 0.425 |
2.784 ± 0.26 | 4.609 ± 0.477 |
5.473 ± 1.625 | 9.65 ± 0.681 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.488 ± 0.194 | 4.033 ± 0.593 |
7.489 ± 1.861 | 3.505 ± 0.323 |
5.377 ± 0.333 | 7.249 ± 1.519 |
6.097 ± 0.164 | 6.673 ± 0.18 |
1.344 ± 0.295 | 3.265 ± 0.509 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
