
Squirrel monkey simian foamy virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Simiispumavirus
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
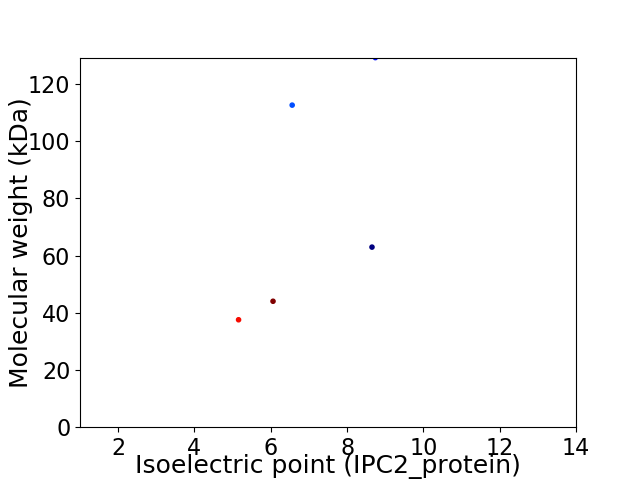
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5JWU9|D5JWU9_9RETR Bel2 OS=Squirrel monkey simian foamy virus OX=2170201 GN=bel2 PE=4 SV=1
MM1 pKa = 7.75ASYY4 pKa = 8.21KK5 pKa = 9.44TQGSVKK11 pKa = 10.28DD12 pKa = 4.02DD13 pKa = 3.53LVSFLQEE20 pKa = 4.26FPLEE24 pKa = 4.47DD25 pKa = 4.84LDD27 pKa = 6.38DD28 pKa = 4.49IPTPAPCGMFEE39 pKa = 4.77EE40 pKa = 5.01YY41 pKa = 10.5FPPTPMDD48 pKa = 3.8PVAMGSEE55 pKa = 4.53YY56 pKa = 11.01NVAPLAEE63 pKa = 5.2LPDD66 pKa = 3.63QLLSVTFPASPASTEE81 pKa = 4.1SSSEE85 pKa = 3.95AGAQDD90 pKa = 3.54YY91 pKa = 9.83TGEE94 pKa = 4.05GTAAGEE100 pKa = 4.47APPEE104 pKa = 4.37AGPSRR109 pKa = 11.84SNSQPGGTPFFWLQEE124 pKa = 3.68ARR126 pKa = 11.84KK127 pKa = 9.96RR128 pKa = 11.84SDD130 pKa = 3.32YY131 pKa = 11.39DD132 pKa = 3.39CWGLNYY138 pKa = 9.02FFPKK142 pKa = 9.6PGQRR146 pKa = 11.84EE147 pKa = 3.96FYY149 pKa = 9.15IQRR152 pKa = 11.84SILTTAGYY160 pKa = 10.43RR161 pKa = 11.84PDD163 pKa = 3.59LLRR166 pKa = 11.84YY167 pKa = 8.71SAAGGWYY174 pKa = 9.36PIILPHH180 pKa = 6.69EE181 pKa = 4.55GVLGNTFQVYY191 pKa = 7.45YY192 pKa = 10.86LCLKK196 pKa = 10.58CGDD199 pKa = 4.35EE200 pKa = 4.24VWDD203 pKa = 3.97PLLYY207 pKa = 10.78LWDD210 pKa = 3.74QDD212 pKa = 4.11LLAFYY217 pKa = 9.81RR218 pKa = 11.84AWEE221 pKa = 4.34TTPHH225 pKa = 5.94TSSPLSVLRR234 pKa = 11.84RR235 pKa = 11.84HH236 pKa = 6.56DD237 pKa = 3.51ASCRR241 pKa = 11.84YY242 pKa = 8.86LHH244 pKa = 6.84HH245 pKa = 6.59NAYY248 pKa = 10.33NYY250 pKa = 9.7SEE252 pKa = 4.44RR253 pKa = 11.84PNIRR257 pKa = 11.84KK258 pKa = 8.82PRR260 pKa = 11.84RR261 pKa = 11.84DD262 pKa = 3.54PGLRR266 pKa = 11.84WCTGKK271 pKa = 10.23RR272 pKa = 11.84RR273 pKa = 11.84LLRR276 pKa = 11.84DD277 pKa = 3.07SCEE280 pKa = 4.13EE281 pKa = 3.89LVHH284 pKa = 6.97PGTLPNPTQWTQLTPSHH301 pKa = 5.89MHH303 pKa = 6.59PLAALSEE310 pKa = 4.18LLQPEE315 pKa = 4.88LQTSPSSPLEE325 pKa = 3.85GTKK328 pKa = 9.89KK329 pKa = 10.42RR330 pKa = 11.84KK331 pKa = 9.12LAEE334 pKa = 3.85
MM1 pKa = 7.75ASYY4 pKa = 8.21KK5 pKa = 9.44TQGSVKK11 pKa = 10.28DD12 pKa = 4.02DD13 pKa = 3.53LVSFLQEE20 pKa = 4.26FPLEE24 pKa = 4.47DD25 pKa = 4.84LDD27 pKa = 6.38DD28 pKa = 4.49IPTPAPCGMFEE39 pKa = 4.77EE40 pKa = 5.01YY41 pKa = 10.5FPPTPMDD48 pKa = 3.8PVAMGSEE55 pKa = 4.53YY56 pKa = 11.01NVAPLAEE63 pKa = 5.2LPDD66 pKa = 3.63QLLSVTFPASPASTEE81 pKa = 4.1SSSEE85 pKa = 3.95AGAQDD90 pKa = 3.54YY91 pKa = 9.83TGEE94 pKa = 4.05GTAAGEE100 pKa = 4.47APPEE104 pKa = 4.37AGPSRR109 pKa = 11.84SNSQPGGTPFFWLQEE124 pKa = 3.68ARR126 pKa = 11.84KK127 pKa = 9.96RR128 pKa = 11.84SDD130 pKa = 3.32YY131 pKa = 11.39DD132 pKa = 3.39CWGLNYY138 pKa = 9.02FFPKK142 pKa = 9.6PGQRR146 pKa = 11.84EE147 pKa = 3.96FYY149 pKa = 9.15IQRR152 pKa = 11.84SILTTAGYY160 pKa = 10.43RR161 pKa = 11.84PDD163 pKa = 3.59LLRR166 pKa = 11.84YY167 pKa = 8.71SAAGGWYY174 pKa = 9.36PIILPHH180 pKa = 6.69EE181 pKa = 4.55GVLGNTFQVYY191 pKa = 7.45YY192 pKa = 10.86LCLKK196 pKa = 10.58CGDD199 pKa = 4.35EE200 pKa = 4.24VWDD203 pKa = 3.97PLLYY207 pKa = 10.78LWDD210 pKa = 3.74QDD212 pKa = 4.11LLAFYY217 pKa = 9.81RR218 pKa = 11.84AWEE221 pKa = 4.34TTPHH225 pKa = 5.94TSSPLSVLRR234 pKa = 11.84RR235 pKa = 11.84HH236 pKa = 6.56DD237 pKa = 3.51ASCRR241 pKa = 11.84YY242 pKa = 8.86LHH244 pKa = 6.84HH245 pKa = 6.59NAYY248 pKa = 10.33NYY250 pKa = 9.7SEE252 pKa = 4.44RR253 pKa = 11.84PNIRR257 pKa = 11.84KK258 pKa = 8.82PRR260 pKa = 11.84RR261 pKa = 11.84DD262 pKa = 3.54PGLRR266 pKa = 11.84WCTGKK271 pKa = 10.23RR272 pKa = 11.84RR273 pKa = 11.84LLRR276 pKa = 11.84DD277 pKa = 3.07SCEE280 pKa = 4.13EE281 pKa = 3.89LVHH284 pKa = 6.97PGTLPNPTQWTQLTPSHH301 pKa = 5.89MHH303 pKa = 6.59PLAALSEE310 pKa = 4.18LLQPEE315 pKa = 4.88LQTSPSSPLEE325 pKa = 3.85GTKK328 pKa = 9.89KK329 pKa = 10.42RR330 pKa = 11.84KK331 pKa = 9.12LAEE334 pKa = 3.85
Molecular weight: 37.58 kDa
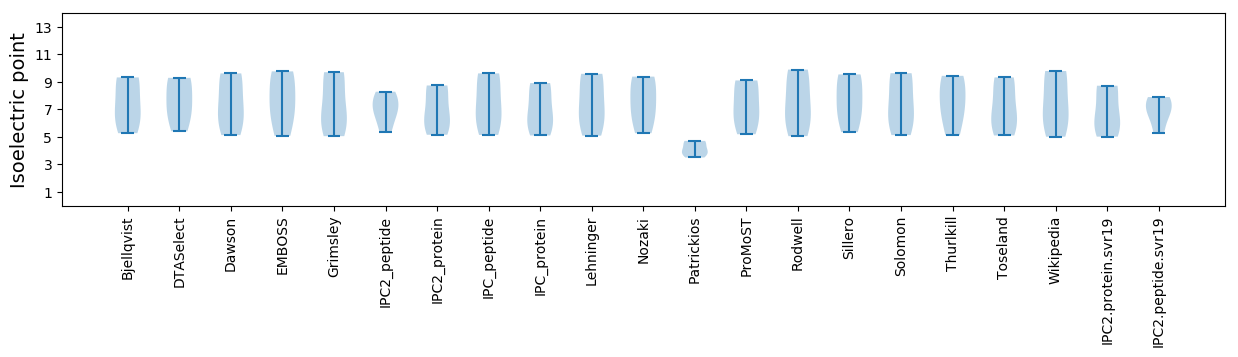
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5JWU7|D5JWU7_9RETR Env leader protein OS=Squirrel monkey simian foamy virus OX=2170201 GN=env PE=4 SV=1
MM1 pKa = 7.07YY2 pKa = 10.27QPQHH6 pKa = 5.04QLQVEE11 pKa = 4.03IHH13 pKa = 5.57DD14 pKa = 4.15QKK16 pKa = 11.51LIGYY20 pKa = 7.56WDD22 pKa = 3.58TGAQITCIPQVYY34 pKa = 9.92LEE36 pKa = 3.98QEE38 pKa = 4.11KK39 pKa = 10.6PIGKK43 pKa = 8.74HH44 pKa = 5.29VIEE47 pKa = 4.39TVNGKK52 pKa = 5.99TQRR55 pKa = 11.84DD56 pKa = 3.48AYY58 pKa = 10.3YY59 pKa = 10.11IKK61 pKa = 10.95LKK63 pKa = 10.84INGKK67 pKa = 9.86KK68 pKa = 9.61IEE70 pKa = 4.29TEE72 pKa = 4.26VIPSPFSYY80 pKa = 11.33ALITPNDD87 pKa = 3.78IPWFKK92 pKa = 10.67PGGIEE97 pKa = 4.09LTEE100 pKa = 4.08KK101 pKa = 11.09LPIQDD106 pKa = 3.68YY107 pKa = 10.68KK108 pKa = 11.86DD109 pKa = 4.32NIVKK113 pKa = 10.43RR114 pKa = 11.84ADD116 pKa = 2.97ITKK119 pKa = 9.66EE120 pKa = 4.04EE121 pKa = 4.09KK122 pKa = 10.86GMLYY126 pKa = 10.73KK127 pKa = 10.92LLDD130 pKa = 4.03KK131 pKa = 11.01YY132 pKa = 11.48DD133 pKa = 4.5PLWQQWEE140 pKa = 4.26NQVGNRR146 pKa = 11.84QITPHH151 pKa = 7.18IIATGTINPKK161 pKa = 8.77PQKK164 pKa = 9.83QYY166 pKa = 10.99HH167 pKa = 6.53INPKK171 pKa = 9.83AKK173 pKa = 9.83PSIQIVINDD182 pKa = 3.98LLKK185 pKa = 10.96QGVLKK190 pKa = 10.16QQNSIMNTPIYY201 pKa = 9.13PVPKK205 pKa = 10.27TEE207 pKa = 4.46GKK209 pKa = 8.74WRR211 pKa = 11.84MVLDD215 pKa = 3.44YY216 pKa = 10.78RR217 pKa = 11.84AVNKK221 pKa = 7.69TIPLIAAQNQHH232 pKa = 5.68SAGILTNLVRR242 pKa = 11.84QKK244 pKa = 10.91YY245 pKa = 9.88KK246 pKa = 10.25STIDD250 pKa = 3.45LSNGFWAHH258 pKa = 7.32PIDD261 pKa = 5.96QDD263 pKa = 4.25SQWITAFTWEE273 pKa = 4.26GKK275 pKa = 9.52QYY277 pKa = 10.6VWTRR281 pKa = 11.84LPQGFLNSPALFTADD296 pKa = 3.46VVDD299 pKa = 4.6LLKK302 pKa = 10.61EE303 pKa = 3.89IPNVNVYY310 pKa = 10.51VDD312 pKa = 4.78DD313 pKa = 4.94IYY315 pKa = 11.87VSTEE319 pKa = 3.76TINQHH324 pKa = 5.31FQVLDD329 pKa = 3.9KK330 pKa = 10.91IFQKK334 pKa = 10.73LLQAGYY340 pKa = 10.27VVSLKK345 pKa = 10.62KK346 pKa = 10.87SNLCRR351 pKa = 11.84YY352 pKa = 7.37EE353 pKa = 3.93VTFLGFTISKK363 pKa = 9.18YY364 pKa = 10.68GRR366 pKa = 11.84GLTEE370 pKa = 3.96EE371 pKa = 4.25FQEE374 pKa = 4.23KK375 pKa = 9.93LRR377 pKa = 11.84NISPPNSLKK386 pKa = 10.33QLQSILGLLNFARR399 pKa = 11.84NFIPNFSEE407 pKa = 4.63LIKK410 pKa = 10.29PLYY413 pKa = 9.8EE414 pKa = 5.22LISTAQGQSISWEE427 pKa = 4.21PKK429 pKa = 9.63HH430 pKa = 6.86SQALNNLIIALNHH443 pKa = 6.72ADD445 pKa = 4.15NLEE448 pKa = 3.94QRR450 pKa = 11.84NGEE453 pKa = 4.06VPLVIKK459 pKa = 10.62INASNTTGYY468 pKa = 10.17IRR470 pKa = 11.84FYY472 pKa = 10.64NKK474 pKa = 9.8NGKK477 pKa = 9.52RR478 pKa = 11.84PIAYY482 pKa = 9.25ASHH485 pKa = 6.18VFNHH489 pKa = 6.15TEE491 pKa = 3.83QKK493 pKa = 8.2FTPVEE498 pKa = 3.96KK499 pKa = 10.77LLTTMHH505 pKa = 6.77KK506 pKa = 10.56AIIKK510 pKa = 10.58GIDD513 pKa = 3.44LAIGQPIEE521 pKa = 4.13IYY523 pKa = 10.67SPIVSMQKK531 pKa = 9.61LQKK534 pKa = 8.97ITLPEE539 pKa = 3.99RR540 pKa = 11.84KK541 pKa = 9.44ALSTRR546 pKa = 11.84WLSWLSYY553 pKa = 10.62IEE555 pKa = 4.34DD556 pKa = 3.54PRR558 pKa = 11.84FLFIYY563 pKa = 10.47DD564 pKa = 3.53KK565 pKa = 10.64TLPDD569 pKa = 4.21LKK571 pKa = 10.75EE572 pKa = 3.99MPPTQTDD579 pKa = 3.84DD580 pKa = 3.98YY581 pKa = 11.5NPMLPLHH588 pKa = 6.26QYY590 pKa = 10.62LAVFYY595 pKa = 10.19TDD597 pKa = 3.35GSSIKK602 pKa = 10.93SPDD605 pKa = 3.23PTKK608 pKa = 9.81THH610 pKa = 6.22SSGMGIVQAIYY621 pKa = 10.41EE622 pKa = 4.31PNFQIKK628 pKa = 8.73HH629 pKa = 4.41QWSIPLGDD637 pKa = 3.74HH638 pKa = 5.83TAQYY642 pKa = 11.48AEE644 pKa = 3.91IAAVEE649 pKa = 4.35FACKK653 pKa = 9.82KK654 pKa = 9.64ALQVTGPVLIVTDD667 pKa = 3.44SDD669 pKa = 4.12YY670 pKa = 11.49VARR673 pKa = 11.84SVNNEE678 pKa = 3.71LNFWRR683 pKa = 11.84SNGFVNNKK691 pKa = 9.32KK692 pKa = 10.41KK693 pKa = 9.96PLKK696 pKa = 10.38HH697 pKa = 5.16ISKK700 pKa = 9.02WKK702 pKa = 10.23SISEE706 pKa = 4.05SLLLHH711 pKa = 6.01KK712 pKa = 10.64NITIVHH718 pKa = 6.06EE719 pKa = 4.78PGHH722 pKa = 5.86QPSSTSVHH730 pKa = 4.78TQGNALADD738 pKa = 3.66KK739 pKa = 10.63LAVQGSYY746 pKa = 10.11TINNITIKK754 pKa = 10.51PSLDD758 pKa = 3.31TEE760 pKa = 4.16LRR762 pKa = 11.84AVLEE766 pKa = 4.45GKK768 pKa = 9.88LPKK771 pKa = 10.33GYY773 pKa = 9.49PKK775 pKa = 10.38NLKK778 pKa = 10.14YY779 pKa = 10.49EE780 pKa = 4.28YY781 pKa = 10.1NSPNLIVIRR790 pKa = 11.84KK791 pKa = 7.34EE792 pKa = 3.8GQRR795 pKa = 11.84IIPPLSDD802 pKa = 3.06RR803 pKa = 11.84PKK805 pKa = 10.33LVKK808 pKa = 9.68QAHH811 pKa = 5.72EE812 pKa = 3.95LAHH815 pKa = 6.27TGRR818 pKa = 11.84EE819 pKa = 4.12ATLLRR824 pKa = 11.84LQNQYY829 pKa = 7.67WWPKK833 pKa = 6.64MRR835 pKa = 11.84KK836 pKa = 9.21DD837 pKa = 3.41VSHH840 pKa = 7.31CLRR843 pKa = 11.84TCMPCLQTNSTNLTTTRR860 pKa = 11.84PFQQIRR866 pKa = 11.84PSKK869 pKa = 10.36PFDD872 pKa = 3.39KK873 pKa = 11.06YY874 pKa = 10.63YY875 pKa = 10.25IDD877 pKa = 4.52YY878 pKa = 10.48IGPLPPSEE886 pKa = 4.4GYY888 pKa = 10.44SYY890 pKa = 11.68VLVVVDD896 pKa = 4.87SATGFCWLYY905 pKa = 8.1PTKK908 pKa = 10.71APSTRR913 pKa = 11.84ATVKK917 pKa = 10.57SLNFLLGIAVPKK929 pKa = 10.15ILHH932 pKa = 6.43SDD934 pKa = 3.04QGSAFTSSDD943 pKa = 3.03FANWAKK949 pKa = 10.45EE950 pKa = 3.91KK951 pKa = 10.64EE952 pKa = 4.05ITLEE956 pKa = 3.94FSTPYY961 pKa = 10.25HH962 pKa = 5.98PQSSGKK968 pKa = 8.29VEE970 pKa = 4.21RR971 pKa = 11.84KK972 pKa = 8.28NQEE975 pKa = 3.51IKK977 pKa = 10.81KK978 pKa = 10.1LLTKK982 pKa = 10.75LLVGRR987 pKa = 11.84PAKK990 pKa = 9.64WYY992 pKa = 8.75PLIPSVQLALNNTYY1006 pKa = 10.44SPKK1009 pKa = 10.24IKK1011 pKa = 9.59LTPHH1015 pKa = 5.61QLLFGVDD1022 pKa = 3.69GNIPFANSDD1031 pKa = 3.58TLDD1034 pKa = 3.48LKK1036 pKa = 11.06RR1037 pKa = 11.84EE1038 pKa = 4.05EE1039 pKa = 4.2EE1040 pKa = 4.07LALLSEE1046 pKa = 4.19IRR1048 pKa = 11.84TTLSTVSPEE1057 pKa = 4.23PFPSTAKK1064 pKa = 9.1TWTPSVGLLVQEE1076 pKa = 4.17RR1077 pKa = 11.84VYY1079 pKa = 10.73RR1080 pKa = 11.84PSQLRR1085 pKa = 11.84PKK1087 pKa = 8.48WKK1089 pKa = 10.0KK1090 pKa = 7.22PTPILEE1096 pKa = 4.32VLNEE1100 pKa = 3.99RR1101 pKa = 11.84TVVIDD1106 pKa = 3.26NNGQRR1111 pKa = 11.84RR1112 pKa = 11.84TVSVDD1117 pKa = 2.81NLKK1120 pKa = 8.82YY1121 pKa = 9.39TPHH1124 pKa = 6.08QKK1126 pKa = 10.64DD1127 pKa = 3.29GEE1129 pKa = 4.71TYY1131 pKa = 10.67DD1132 pKa = 3.59SSS1134 pKa = 3.64
MM1 pKa = 7.07YY2 pKa = 10.27QPQHH6 pKa = 5.04QLQVEE11 pKa = 4.03IHH13 pKa = 5.57DD14 pKa = 4.15QKK16 pKa = 11.51LIGYY20 pKa = 7.56WDD22 pKa = 3.58TGAQITCIPQVYY34 pKa = 9.92LEE36 pKa = 3.98QEE38 pKa = 4.11KK39 pKa = 10.6PIGKK43 pKa = 8.74HH44 pKa = 5.29VIEE47 pKa = 4.39TVNGKK52 pKa = 5.99TQRR55 pKa = 11.84DD56 pKa = 3.48AYY58 pKa = 10.3YY59 pKa = 10.11IKK61 pKa = 10.95LKK63 pKa = 10.84INGKK67 pKa = 9.86KK68 pKa = 9.61IEE70 pKa = 4.29TEE72 pKa = 4.26VIPSPFSYY80 pKa = 11.33ALITPNDD87 pKa = 3.78IPWFKK92 pKa = 10.67PGGIEE97 pKa = 4.09LTEE100 pKa = 4.08KK101 pKa = 11.09LPIQDD106 pKa = 3.68YY107 pKa = 10.68KK108 pKa = 11.86DD109 pKa = 4.32NIVKK113 pKa = 10.43RR114 pKa = 11.84ADD116 pKa = 2.97ITKK119 pKa = 9.66EE120 pKa = 4.04EE121 pKa = 4.09KK122 pKa = 10.86GMLYY126 pKa = 10.73KK127 pKa = 10.92LLDD130 pKa = 4.03KK131 pKa = 11.01YY132 pKa = 11.48DD133 pKa = 4.5PLWQQWEE140 pKa = 4.26NQVGNRR146 pKa = 11.84QITPHH151 pKa = 7.18IIATGTINPKK161 pKa = 8.77PQKK164 pKa = 9.83QYY166 pKa = 10.99HH167 pKa = 6.53INPKK171 pKa = 9.83AKK173 pKa = 9.83PSIQIVINDD182 pKa = 3.98LLKK185 pKa = 10.96QGVLKK190 pKa = 10.16QQNSIMNTPIYY201 pKa = 9.13PVPKK205 pKa = 10.27TEE207 pKa = 4.46GKK209 pKa = 8.74WRR211 pKa = 11.84MVLDD215 pKa = 3.44YY216 pKa = 10.78RR217 pKa = 11.84AVNKK221 pKa = 7.69TIPLIAAQNQHH232 pKa = 5.68SAGILTNLVRR242 pKa = 11.84QKK244 pKa = 10.91YY245 pKa = 9.88KK246 pKa = 10.25STIDD250 pKa = 3.45LSNGFWAHH258 pKa = 7.32PIDD261 pKa = 5.96QDD263 pKa = 4.25SQWITAFTWEE273 pKa = 4.26GKK275 pKa = 9.52QYY277 pKa = 10.6VWTRR281 pKa = 11.84LPQGFLNSPALFTADD296 pKa = 3.46VVDD299 pKa = 4.6LLKK302 pKa = 10.61EE303 pKa = 3.89IPNVNVYY310 pKa = 10.51VDD312 pKa = 4.78DD313 pKa = 4.94IYY315 pKa = 11.87VSTEE319 pKa = 3.76TINQHH324 pKa = 5.31FQVLDD329 pKa = 3.9KK330 pKa = 10.91IFQKK334 pKa = 10.73LLQAGYY340 pKa = 10.27VVSLKK345 pKa = 10.62KK346 pKa = 10.87SNLCRR351 pKa = 11.84YY352 pKa = 7.37EE353 pKa = 3.93VTFLGFTISKK363 pKa = 9.18YY364 pKa = 10.68GRR366 pKa = 11.84GLTEE370 pKa = 3.96EE371 pKa = 4.25FQEE374 pKa = 4.23KK375 pKa = 9.93LRR377 pKa = 11.84NISPPNSLKK386 pKa = 10.33QLQSILGLLNFARR399 pKa = 11.84NFIPNFSEE407 pKa = 4.63LIKK410 pKa = 10.29PLYY413 pKa = 9.8EE414 pKa = 5.22LISTAQGQSISWEE427 pKa = 4.21PKK429 pKa = 9.63HH430 pKa = 6.86SQALNNLIIALNHH443 pKa = 6.72ADD445 pKa = 4.15NLEE448 pKa = 3.94QRR450 pKa = 11.84NGEE453 pKa = 4.06VPLVIKK459 pKa = 10.62INASNTTGYY468 pKa = 10.17IRR470 pKa = 11.84FYY472 pKa = 10.64NKK474 pKa = 9.8NGKK477 pKa = 9.52RR478 pKa = 11.84PIAYY482 pKa = 9.25ASHH485 pKa = 6.18VFNHH489 pKa = 6.15TEE491 pKa = 3.83QKK493 pKa = 8.2FTPVEE498 pKa = 3.96KK499 pKa = 10.77LLTTMHH505 pKa = 6.77KK506 pKa = 10.56AIIKK510 pKa = 10.58GIDD513 pKa = 3.44LAIGQPIEE521 pKa = 4.13IYY523 pKa = 10.67SPIVSMQKK531 pKa = 9.61LQKK534 pKa = 8.97ITLPEE539 pKa = 3.99RR540 pKa = 11.84KK541 pKa = 9.44ALSTRR546 pKa = 11.84WLSWLSYY553 pKa = 10.62IEE555 pKa = 4.34DD556 pKa = 3.54PRR558 pKa = 11.84FLFIYY563 pKa = 10.47DD564 pKa = 3.53KK565 pKa = 10.64TLPDD569 pKa = 4.21LKK571 pKa = 10.75EE572 pKa = 3.99MPPTQTDD579 pKa = 3.84DD580 pKa = 3.98YY581 pKa = 11.5NPMLPLHH588 pKa = 6.26QYY590 pKa = 10.62LAVFYY595 pKa = 10.19TDD597 pKa = 3.35GSSIKK602 pKa = 10.93SPDD605 pKa = 3.23PTKK608 pKa = 9.81THH610 pKa = 6.22SSGMGIVQAIYY621 pKa = 10.41EE622 pKa = 4.31PNFQIKK628 pKa = 8.73HH629 pKa = 4.41QWSIPLGDD637 pKa = 3.74HH638 pKa = 5.83TAQYY642 pKa = 11.48AEE644 pKa = 3.91IAAVEE649 pKa = 4.35FACKK653 pKa = 9.82KK654 pKa = 9.64ALQVTGPVLIVTDD667 pKa = 3.44SDD669 pKa = 4.12YY670 pKa = 11.49VARR673 pKa = 11.84SVNNEE678 pKa = 3.71LNFWRR683 pKa = 11.84SNGFVNNKK691 pKa = 9.32KK692 pKa = 10.41KK693 pKa = 9.96PLKK696 pKa = 10.38HH697 pKa = 5.16ISKK700 pKa = 9.02WKK702 pKa = 10.23SISEE706 pKa = 4.05SLLLHH711 pKa = 6.01KK712 pKa = 10.64NITIVHH718 pKa = 6.06EE719 pKa = 4.78PGHH722 pKa = 5.86QPSSTSVHH730 pKa = 4.78TQGNALADD738 pKa = 3.66KK739 pKa = 10.63LAVQGSYY746 pKa = 10.11TINNITIKK754 pKa = 10.51PSLDD758 pKa = 3.31TEE760 pKa = 4.16LRR762 pKa = 11.84AVLEE766 pKa = 4.45GKK768 pKa = 9.88LPKK771 pKa = 10.33GYY773 pKa = 9.49PKK775 pKa = 10.38NLKK778 pKa = 10.14YY779 pKa = 10.49EE780 pKa = 4.28YY781 pKa = 10.1NSPNLIVIRR790 pKa = 11.84KK791 pKa = 7.34EE792 pKa = 3.8GQRR795 pKa = 11.84IIPPLSDD802 pKa = 3.06RR803 pKa = 11.84PKK805 pKa = 10.33LVKK808 pKa = 9.68QAHH811 pKa = 5.72EE812 pKa = 3.95LAHH815 pKa = 6.27TGRR818 pKa = 11.84EE819 pKa = 4.12ATLLRR824 pKa = 11.84LQNQYY829 pKa = 7.67WWPKK833 pKa = 6.64MRR835 pKa = 11.84KK836 pKa = 9.21DD837 pKa = 3.41VSHH840 pKa = 7.31CLRR843 pKa = 11.84TCMPCLQTNSTNLTTTRR860 pKa = 11.84PFQQIRR866 pKa = 11.84PSKK869 pKa = 10.36PFDD872 pKa = 3.39KK873 pKa = 11.06YY874 pKa = 10.63YY875 pKa = 10.25IDD877 pKa = 4.52YY878 pKa = 10.48IGPLPPSEE886 pKa = 4.4GYY888 pKa = 10.44SYY890 pKa = 11.68VLVVVDD896 pKa = 4.87SATGFCWLYY905 pKa = 8.1PTKK908 pKa = 10.71APSTRR913 pKa = 11.84ATVKK917 pKa = 10.57SLNFLLGIAVPKK929 pKa = 10.15ILHH932 pKa = 6.43SDD934 pKa = 3.04QGSAFTSSDD943 pKa = 3.03FANWAKK949 pKa = 10.45EE950 pKa = 3.91KK951 pKa = 10.64EE952 pKa = 4.05ITLEE956 pKa = 3.94FSTPYY961 pKa = 10.25HH962 pKa = 5.98PQSSGKK968 pKa = 8.29VEE970 pKa = 4.21RR971 pKa = 11.84KK972 pKa = 8.28NQEE975 pKa = 3.51IKK977 pKa = 10.81KK978 pKa = 10.1LLTKK982 pKa = 10.75LLVGRR987 pKa = 11.84PAKK990 pKa = 9.64WYY992 pKa = 8.75PLIPSVQLALNNTYY1006 pKa = 10.44SPKK1009 pKa = 10.24IKK1011 pKa = 9.59LTPHH1015 pKa = 5.61QLLFGVDD1022 pKa = 3.69GNIPFANSDD1031 pKa = 3.58TLDD1034 pKa = 3.48LKK1036 pKa = 11.06RR1037 pKa = 11.84EE1038 pKa = 4.05EE1039 pKa = 4.2EE1040 pKa = 4.07LALLSEE1046 pKa = 4.19IRR1048 pKa = 11.84TTLSTVSPEE1057 pKa = 4.23PFPSTAKK1064 pKa = 9.1TWTPSVGLLVQEE1076 pKa = 4.17RR1077 pKa = 11.84VYY1079 pKa = 10.73RR1080 pKa = 11.84PSQLRR1085 pKa = 11.84PKK1087 pKa = 8.48WKK1089 pKa = 10.0KK1090 pKa = 7.22PTPILEE1096 pKa = 4.32VLNEE1100 pKa = 3.99RR1101 pKa = 11.84TVVIDD1106 pKa = 3.26NNGQRR1111 pKa = 11.84RR1112 pKa = 11.84TVSVDD1117 pKa = 2.81NLKK1120 pKa = 8.82YY1121 pKa = 9.39TPHH1124 pKa = 6.08QKK1126 pKa = 10.64DD1127 pKa = 3.29GEE1129 pKa = 4.71TYY1131 pKa = 10.67DD1132 pKa = 3.59SSS1134 pKa = 3.64
Molecular weight: 129.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3399 |
334 |
1134 |
679.8 |
77.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.707 ± 0.393 | 1.618 ± 0.482 |
4.237 ± 0.254 | 5.855 ± 0.519 |
3.001 ± 0.173 | 5.737 ± 0.562 |
2.354 ± 0.211 | 6.884 ± 0.811 |
6.002 ± 1.076 | 9.768 ± 0.456 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.618 ± 0.269 | 5.296 ± 0.709 |
7.502 ± 0.787 | 5.355 ± 0.288 |
4.766 ± 0.702 | 6.737 ± 0.328 |
6.826 ± 0.248 | 5.325 ± 0.298 |
2.295 ± 0.293 | 4.119 ± 0.291 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
