
Fragaria chiloensis latent virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins
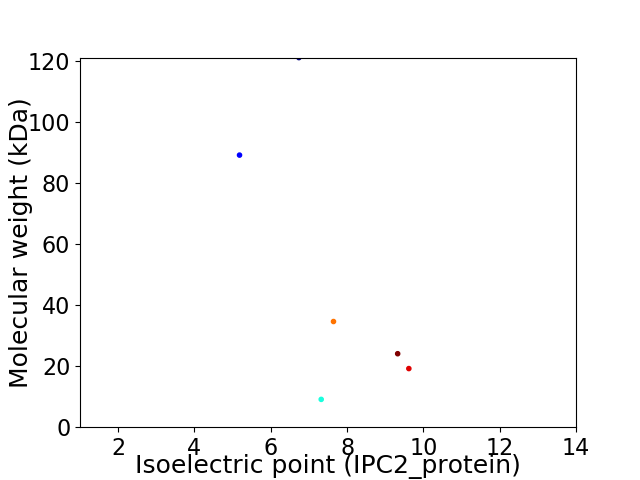
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q5VI74|Q5VI74_9BROM RNA-directed RNA polymerase 2a OS=Fragaria chiloensis latent virus OX=255238 PE=4 SV=1
MM1 pKa = 7.69FCPKK5 pKa = 9.76ILSFVPSSEE14 pKa = 3.72PWVGSEE20 pKa = 4.86SLDD23 pKa = 3.46ASGDD27 pKa = 3.35IPTIDD32 pKa = 3.87DD33 pKa = 3.9WYY35 pKa = 11.3LLTNTVLAEE44 pKa = 3.74RR45 pKa = 11.84LVIEE49 pKa = 5.27RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84QLEE55 pKa = 4.25KK56 pKa = 11.08VVGQYY61 pKa = 7.72KK62 pKa = 8.97TLLHH66 pKa = 5.71FVGWCYY72 pKa = 10.91LEE74 pKa = 3.91QVVFKK79 pKa = 9.46TSKK82 pKa = 10.74YY83 pKa = 9.34KK84 pKa = 11.23GEE86 pKa = 4.21FNTFPFLVEE95 pKa = 3.73RR96 pKa = 11.84FFAAFDD102 pKa = 3.92LEE104 pKa = 4.31APQYY108 pKa = 10.47EE109 pKa = 4.75VLPMQSDD116 pKa = 4.15CLSDD120 pKa = 4.41FFDD123 pKa = 3.95VGCVDD128 pKa = 4.43LPLPIEE134 pKa = 4.61EE135 pKa = 4.81DD136 pKa = 4.06TISLPSDD143 pKa = 3.13FHH145 pKa = 8.61RR146 pKa = 11.84SISSLSYY153 pKa = 9.01PSCGSSSMEE162 pKa = 4.0SGRR165 pKa = 11.84TSSGGPEE172 pKa = 4.01STSSDD177 pKa = 3.7VCHH180 pKa = 7.04EE181 pKa = 4.45IYY183 pKa = 10.63NAGPVDD189 pKa = 4.25PMPQCSRR196 pKa = 11.84RR197 pKa = 11.84WSDD200 pKa = 3.34YY201 pKa = 9.53TSEE204 pKa = 4.05EE205 pKa = 4.46DD206 pKa = 3.34EE207 pKa = 5.01CEE209 pKa = 4.02LVPLSGDD216 pKa = 3.4TLPRR220 pKa = 11.84GEE222 pKa = 4.58IPNEE226 pKa = 3.87IVSVSATVNRR236 pKa = 11.84CLEE239 pKa = 4.06SSYY242 pKa = 10.76MSTDD246 pKa = 2.88IPDD249 pKa = 4.03LGRR252 pKa = 11.84FIEE255 pKa = 4.58EE256 pKa = 3.83PVEE259 pKa = 3.79FDD261 pKa = 3.11GFDD264 pKa = 2.89RR265 pKa = 11.84VVAVNDD271 pKa = 2.92WWSGMKK277 pKa = 10.15LVPNLGIPDD286 pKa = 4.29DD287 pKa = 4.4SDD289 pKa = 4.39VIFSGILSSAPEE301 pKa = 4.15VMQSAIDD308 pKa = 3.64EE309 pKa = 4.53FLPLHH314 pKa = 5.96HH315 pKa = 6.91QIDD318 pKa = 3.64DD319 pKa = 3.71RR320 pKa = 11.84FFQEE324 pKa = 3.88IVEE327 pKa = 4.31TSDD330 pKa = 3.8ISLEE334 pKa = 4.12LEE336 pKa = 3.98NCSFDD341 pKa = 4.01LSSFKK346 pKa = 10.5SWSSHH351 pKa = 5.29EE352 pKa = 4.49SGCCSSLNSGMTSTRR367 pKa = 11.84TNTFRR372 pKa = 11.84EE373 pKa = 3.74AALAIKK379 pKa = 9.01KK380 pKa = 10.45RR381 pKa = 11.84NMNVPAISSSCDD393 pKa = 2.8IGKK396 pKa = 10.1VSDD399 pKa = 3.94DD400 pKa = 3.53VVEE403 pKa = 4.44KK404 pKa = 10.49FFSRR408 pKa = 11.84IIDD411 pKa = 3.45VDD413 pKa = 3.83KK414 pKa = 11.42LIGLPIIGHH423 pKa = 6.41GEE425 pKa = 3.95LAWFADD431 pKa = 3.89YY432 pKa = 11.43LKK434 pKa = 10.63GKK436 pKa = 8.77PVNEE440 pKa = 4.5DD441 pKa = 3.23MFVDD445 pKa = 5.4PICLVSMDD453 pKa = 4.64KK454 pKa = 10.37YY455 pKa = 9.77RR456 pKa = 11.84HH457 pKa = 4.78MVKK460 pKa = 10.12SQLKK464 pKa = 8.47PVEE467 pKa = 4.55DD468 pKa = 3.5NSLAFEE474 pKa = 4.6RR475 pKa = 11.84PLPATITYY483 pKa = 9.28HH484 pKa = 7.58DD485 pKa = 4.21KK486 pKa = 11.19GKK488 pKa = 10.99VMSTSPLFLALMNRR502 pKa = 11.84LLMSLKK508 pKa = 10.5SKK510 pKa = 10.65ISIPTGKK517 pKa = 9.79FHH519 pKa = 7.59QLFSLDD525 pKa = 3.42AQVFDD530 pKa = 5.98SVLEE534 pKa = 3.99WKK536 pKa = 10.42EE537 pKa = 3.43IDD539 pKa = 3.52FSKK542 pKa = 10.58FDD544 pKa = 3.41KK545 pKa = 10.84SQQEE549 pKa = 4.02LHH551 pKa = 6.93HH552 pKa = 6.2EE553 pKa = 4.26VQKK556 pKa = 11.13KK557 pKa = 9.36IFLRR561 pKa = 11.84LGLPRR566 pKa = 11.84DD567 pKa = 4.16FCDD570 pKa = 2.6TWFTSHH576 pKa = 5.73VRR578 pKa = 11.84SHH580 pKa = 6.86ISDD583 pKa = 3.63PSGLRR588 pKa = 11.84FSVNFQRR595 pKa = 11.84RR596 pKa = 11.84TGDD599 pKa = 2.95AVTYY603 pKa = 10.41LGNTVVTLAVLSYY616 pKa = 11.04VYY618 pKa = 10.58DD619 pKa = 4.05LSDD622 pKa = 4.17PNVLMVVASGDD633 pKa = 3.49DD634 pKa = 3.66SLIGSYY640 pKa = 10.41RR641 pKa = 11.84PLDD644 pKa = 3.48RR645 pKa = 11.84SRR647 pKa = 11.84EE648 pKa = 4.22HH649 pKa = 6.87LCSTLFNFEE658 pKa = 4.24AKK660 pKa = 10.1FPHH663 pKa = 5.93NQPFICSKK671 pKa = 10.66FLLTMPTKK679 pKa = 9.87SGGRR683 pKa = 11.84RR684 pKa = 11.84VVAVPNPLKK693 pKa = 10.83LLIKK697 pKa = 10.55LGIRR701 pKa = 11.84NLQEE705 pKa = 3.97DD706 pKa = 4.47QFDD709 pKa = 3.0AWYY712 pKa = 8.69TSWIDD717 pKa = 3.8LIHH720 pKa = 6.2YY721 pKa = 9.51FNDD724 pKa = 2.8EE725 pKa = 3.98HH726 pKa = 7.66LISVVAEE733 pKa = 4.09MCSYY737 pKa = 10.92RR738 pKa = 11.84YY739 pKa = 9.58LRR741 pKa = 11.84KK742 pKa = 9.37PSMFLKK748 pKa = 10.59PAMCSFNNVFANKK761 pKa = 8.24TKK763 pKa = 10.52LMKK766 pKa = 10.45FLFPSMVLRR775 pKa = 11.84KK776 pKa = 9.77DD777 pKa = 3.03KK778 pKa = 10.81RR779 pKa = 11.84KK780 pKa = 9.88RR781 pKa = 11.84PPRR784 pKa = 11.84KK785 pKa = 9.33RR786 pKa = 3.3
MM1 pKa = 7.69FCPKK5 pKa = 9.76ILSFVPSSEE14 pKa = 3.72PWVGSEE20 pKa = 4.86SLDD23 pKa = 3.46ASGDD27 pKa = 3.35IPTIDD32 pKa = 3.87DD33 pKa = 3.9WYY35 pKa = 11.3LLTNTVLAEE44 pKa = 3.74RR45 pKa = 11.84LVIEE49 pKa = 5.27RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84QLEE55 pKa = 4.25KK56 pKa = 11.08VVGQYY61 pKa = 7.72KK62 pKa = 8.97TLLHH66 pKa = 5.71FVGWCYY72 pKa = 10.91LEE74 pKa = 3.91QVVFKK79 pKa = 9.46TSKK82 pKa = 10.74YY83 pKa = 9.34KK84 pKa = 11.23GEE86 pKa = 4.21FNTFPFLVEE95 pKa = 3.73RR96 pKa = 11.84FFAAFDD102 pKa = 3.92LEE104 pKa = 4.31APQYY108 pKa = 10.47EE109 pKa = 4.75VLPMQSDD116 pKa = 4.15CLSDD120 pKa = 4.41FFDD123 pKa = 3.95VGCVDD128 pKa = 4.43LPLPIEE134 pKa = 4.61EE135 pKa = 4.81DD136 pKa = 4.06TISLPSDD143 pKa = 3.13FHH145 pKa = 8.61RR146 pKa = 11.84SISSLSYY153 pKa = 9.01PSCGSSSMEE162 pKa = 4.0SGRR165 pKa = 11.84TSSGGPEE172 pKa = 4.01STSSDD177 pKa = 3.7VCHH180 pKa = 7.04EE181 pKa = 4.45IYY183 pKa = 10.63NAGPVDD189 pKa = 4.25PMPQCSRR196 pKa = 11.84RR197 pKa = 11.84WSDD200 pKa = 3.34YY201 pKa = 9.53TSEE204 pKa = 4.05EE205 pKa = 4.46DD206 pKa = 3.34EE207 pKa = 5.01CEE209 pKa = 4.02LVPLSGDD216 pKa = 3.4TLPRR220 pKa = 11.84GEE222 pKa = 4.58IPNEE226 pKa = 3.87IVSVSATVNRR236 pKa = 11.84CLEE239 pKa = 4.06SSYY242 pKa = 10.76MSTDD246 pKa = 2.88IPDD249 pKa = 4.03LGRR252 pKa = 11.84FIEE255 pKa = 4.58EE256 pKa = 3.83PVEE259 pKa = 3.79FDD261 pKa = 3.11GFDD264 pKa = 2.89RR265 pKa = 11.84VVAVNDD271 pKa = 2.92WWSGMKK277 pKa = 10.15LVPNLGIPDD286 pKa = 4.29DD287 pKa = 4.4SDD289 pKa = 4.39VIFSGILSSAPEE301 pKa = 4.15VMQSAIDD308 pKa = 3.64EE309 pKa = 4.53FLPLHH314 pKa = 5.96HH315 pKa = 6.91QIDD318 pKa = 3.64DD319 pKa = 3.71RR320 pKa = 11.84FFQEE324 pKa = 3.88IVEE327 pKa = 4.31TSDD330 pKa = 3.8ISLEE334 pKa = 4.12LEE336 pKa = 3.98NCSFDD341 pKa = 4.01LSSFKK346 pKa = 10.5SWSSHH351 pKa = 5.29EE352 pKa = 4.49SGCCSSLNSGMTSTRR367 pKa = 11.84TNTFRR372 pKa = 11.84EE373 pKa = 3.74AALAIKK379 pKa = 9.01KK380 pKa = 10.45RR381 pKa = 11.84NMNVPAISSSCDD393 pKa = 2.8IGKK396 pKa = 10.1VSDD399 pKa = 3.94DD400 pKa = 3.53VVEE403 pKa = 4.44KK404 pKa = 10.49FFSRR408 pKa = 11.84IIDD411 pKa = 3.45VDD413 pKa = 3.83KK414 pKa = 11.42LIGLPIIGHH423 pKa = 6.41GEE425 pKa = 3.95LAWFADD431 pKa = 3.89YY432 pKa = 11.43LKK434 pKa = 10.63GKK436 pKa = 8.77PVNEE440 pKa = 4.5DD441 pKa = 3.23MFVDD445 pKa = 5.4PICLVSMDD453 pKa = 4.64KK454 pKa = 10.37YY455 pKa = 9.77RR456 pKa = 11.84HH457 pKa = 4.78MVKK460 pKa = 10.12SQLKK464 pKa = 8.47PVEE467 pKa = 4.55DD468 pKa = 3.5NSLAFEE474 pKa = 4.6RR475 pKa = 11.84PLPATITYY483 pKa = 9.28HH484 pKa = 7.58DD485 pKa = 4.21KK486 pKa = 11.19GKK488 pKa = 10.99VMSTSPLFLALMNRR502 pKa = 11.84LLMSLKK508 pKa = 10.5SKK510 pKa = 10.65ISIPTGKK517 pKa = 9.79FHH519 pKa = 7.59QLFSLDD525 pKa = 3.42AQVFDD530 pKa = 5.98SVLEE534 pKa = 3.99WKK536 pKa = 10.42EE537 pKa = 3.43IDD539 pKa = 3.52FSKK542 pKa = 10.58FDD544 pKa = 3.41KK545 pKa = 10.84SQQEE549 pKa = 4.02LHH551 pKa = 6.93HH552 pKa = 6.2EE553 pKa = 4.26VQKK556 pKa = 11.13KK557 pKa = 9.36IFLRR561 pKa = 11.84LGLPRR566 pKa = 11.84DD567 pKa = 4.16FCDD570 pKa = 2.6TWFTSHH576 pKa = 5.73VRR578 pKa = 11.84SHH580 pKa = 6.86ISDD583 pKa = 3.63PSGLRR588 pKa = 11.84FSVNFQRR595 pKa = 11.84RR596 pKa = 11.84TGDD599 pKa = 2.95AVTYY603 pKa = 10.41LGNTVVTLAVLSYY616 pKa = 11.04VYY618 pKa = 10.58DD619 pKa = 4.05LSDD622 pKa = 4.17PNVLMVVASGDD633 pKa = 3.49DD634 pKa = 3.66SLIGSYY640 pKa = 10.41RR641 pKa = 11.84PLDD644 pKa = 3.48RR645 pKa = 11.84SRR647 pKa = 11.84EE648 pKa = 4.22HH649 pKa = 6.87LCSTLFNFEE658 pKa = 4.24AKK660 pKa = 10.1FPHH663 pKa = 5.93NQPFICSKK671 pKa = 10.66FLLTMPTKK679 pKa = 9.87SGGRR683 pKa = 11.84RR684 pKa = 11.84VVAVPNPLKK693 pKa = 10.83LLIKK697 pKa = 10.55LGIRR701 pKa = 11.84NLQEE705 pKa = 3.97DD706 pKa = 4.47QFDD709 pKa = 3.0AWYY712 pKa = 8.69TSWIDD717 pKa = 3.8LIHH720 pKa = 6.2YY721 pKa = 9.51FNDD724 pKa = 2.8EE725 pKa = 3.98HH726 pKa = 7.66LISVVAEE733 pKa = 4.09MCSYY737 pKa = 10.92RR738 pKa = 11.84YY739 pKa = 9.58LRR741 pKa = 11.84KK742 pKa = 9.37PSMFLKK748 pKa = 10.59PAMCSFNNVFANKK761 pKa = 8.24TKK763 pKa = 10.52LMKK766 pKa = 10.45FLFPSMVLRR775 pKa = 11.84KK776 pKa = 9.77DD777 pKa = 3.03KK778 pKa = 10.81RR779 pKa = 11.84KK780 pKa = 9.88RR781 pKa = 11.84PPRR784 pKa = 11.84KK785 pKa = 9.33RR786 pKa = 3.3
Molecular weight: 89.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5QDM5|Q5QDM5_9BROM ATP-dependent helicase OS=Fragaria chiloensis latent virus OX=255238 PE=3 SV=1
MM1 pKa = 7.96RR2 pKa = 11.84YY3 pKa = 9.02CRR5 pKa = 11.84CSLIVCRR12 pKa = 11.84ISSMSVVLIYY22 pKa = 10.99LFLSKK27 pKa = 10.44RR28 pKa = 11.84IPSVCLPIFIEE39 pKa = 4.15VFLRR43 pKa = 11.84CLTRR47 pKa = 11.84HH48 pKa = 6.33AVRR51 pKa = 11.84QAWKK55 pKa = 8.85VGVPVRR61 pKa = 11.84EE62 pKa = 4.29VPRR65 pKa = 11.84VRR67 pKa = 11.84PVMFVTKK74 pKa = 10.3FIMPDD79 pKa = 3.12LLIRR83 pKa = 11.84CLSVPVAGQIIPARR97 pKa = 11.84KK98 pKa = 7.94MSVNLFHH105 pKa = 7.36LAVTPYY111 pKa = 10.62RR112 pKa = 11.84EE113 pKa = 4.06VRR115 pKa = 11.84SQMRR119 pKa = 11.84SLALVLRR126 pKa = 11.84SIDD129 pKa = 2.87AWRR132 pKa = 11.84VLICLPIFPILDD144 pKa = 3.91DD145 pKa = 3.98LSKK148 pKa = 11.13NRR150 pKa = 11.84SNLMDD155 pKa = 3.46STVWLLSMTGGLGG168 pKa = 3.17
MM1 pKa = 7.96RR2 pKa = 11.84YY3 pKa = 9.02CRR5 pKa = 11.84CSLIVCRR12 pKa = 11.84ISSMSVVLIYY22 pKa = 10.99LFLSKK27 pKa = 10.44RR28 pKa = 11.84IPSVCLPIFIEE39 pKa = 4.15VFLRR43 pKa = 11.84CLTRR47 pKa = 11.84HH48 pKa = 6.33AVRR51 pKa = 11.84QAWKK55 pKa = 8.85VGVPVRR61 pKa = 11.84EE62 pKa = 4.29VPRR65 pKa = 11.84VRR67 pKa = 11.84PVMFVTKK74 pKa = 10.3FIMPDD79 pKa = 3.12LLIRR83 pKa = 11.84CLSVPVAGQIIPARR97 pKa = 11.84KK98 pKa = 7.94MSVNLFHH105 pKa = 7.36LAVTPYY111 pKa = 10.62RR112 pKa = 11.84EE113 pKa = 4.06VRR115 pKa = 11.84SQMRR119 pKa = 11.84SLALVLRR126 pKa = 11.84SIDD129 pKa = 2.87AWRR132 pKa = 11.84VLICLPIFPILDD144 pKa = 3.91DD145 pKa = 3.98LSKK148 pKa = 11.13NRR150 pKa = 11.84SNLMDD155 pKa = 3.46STVWLLSMTGGLGG168 pKa = 3.17
Molecular weight: 19.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2648 |
78 |
1083 |
441.3 |
49.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.382 ± 1.113 | 2.644 ± 0.332 |
6.156 ± 0.632 | 5.211 ± 0.527 |
5.249 ± 0.474 | 5.211 ± 0.587 |
2.228 ± 0.324 | 5.74 ± 0.489 |
5.098 ± 0.818 | 8.573 ± 0.762 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.719 ± 0.185 | 3.776 ± 0.389 |
5.098 ± 0.641 | 2.53 ± 0.26 |
6.533 ± 1.074 | 8.988 ± 1.302 |
5.816 ± 0.702 | 8.119 ± 0.665 |
1.133 ± 0.169 | 2.795 ± 0.312 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
