
Mesorhizobium ephedrae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
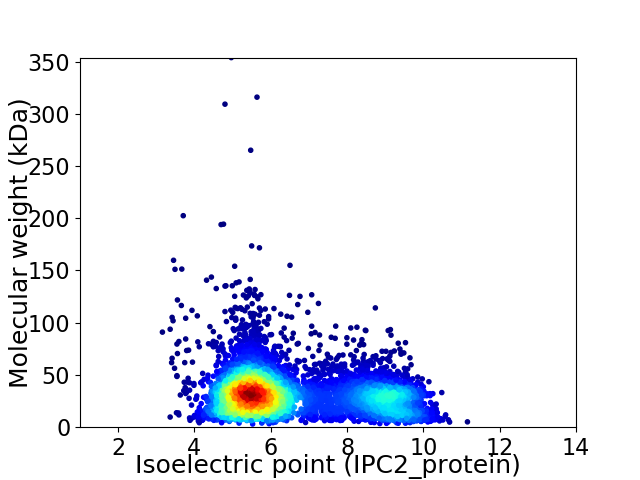
Virtual 2D-PAGE plot for 5676 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P7SS06|A0A2P7SS06_9RHIZ Maleylacetate reductase OS=Mesorhizobium ephedrae OX=2116701 GN=C7I84_02655 PE=4 SV=1
MM1 pKa = 8.34CMMCQTVSRR10 pKa = 11.84FASEE14 pKa = 4.83EE15 pKa = 3.39IDD17 pKa = 3.51APMPSGSVSVAPPQFDD33 pKa = 3.58QMRR36 pKa = 11.84PPYY39 pKa = 10.63DD40 pKa = 3.27MRR42 pKa = 11.84EE43 pKa = 4.01QEE45 pKa = 4.38PGDD48 pKa = 3.55AYY50 pKa = 10.77GAGRR54 pKa = 11.84TALISQVFDD63 pKa = 3.65AADD66 pKa = 3.21GRR68 pKa = 11.84YY69 pKa = 9.17EE70 pKa = 3.81FSGNQDD76 pKa = 1.95IDD78 pKa = 3.3ATLIGSRR85 pKa = 11.84WTLSNLTFSFPTSGTFYY102 pKa = 11.06ADD104 pKa = 2.81EE105 pKa = 5.3GYY107 pKa = 9.37PAGTEE112 pKa = 4.04PAEE115 pKa = 3.9HH116 pKa = 6.07VAFNAMQQDD125 pKa = 3.7AVRR128 pKa = 11.84YY129 pKa = 8.95ALSLVQSYY137 pKa = 9.71TNLTFTEE144 pKa = 4.53VAEE147 pKa = 4.47SATTHH152 pKa = 6.11AEE154 pKa = 3.8LRR156 pKa = 11.84FSQTSYY162 pKa = 11.71DD163 pKa = 4.2DD164 pKa = 3.8VPSAYY169 pKa = 10.38ANFPSSSEE177 pKa = 3.79QAGDD181 pKa = 3.08VWFGMTGQPFYY192 pKa = 10.0DD193 pKa = 4.08TPAMGNWGMATILHH207 pKa = 6.74EE208 pKa = 4.78IGHH211 pKa = 6.45ALGLKK216 pKa = 9.85HH217 pKa = 6.41GHH219 pKa = 5.78QDD221 pKa = 3.35YY222 pKa = 8.54TAVDD226 pKa = 3.99LAGEE230 pKa = 5.19GYY232 pKa = 10.68LDD234 pKa = 3.99PPGGGGPRR242 pKa = 11.84YY243 pKa = 10.14GSAEE247 pKa = 3.92LPANHH252 pKa = 7.57DD253 pKa = 3.76GQDD256 pKa = 3.41WSLMTYY262 pKa = 10.25RR263 pKa = 11.84SDD265 pKa = 3.25PGAPVAFQGEE275 pKa = 5.11GFNQPQTYY283 pKa = 7.44MQNDD287 pKa = 3.39IAALQYY293 pKa = 11.06LYY295 pKa = 11.07GANFDD300 pKa = 4.42TNSSATVYY308 pKa = 10.17SWNPTTGEE316 pKa = 3.81MSIDD320 pKa = 3.58GVGQGAPTSNKK331 pKa = 9.63ISMTLWDD338 pKa = 4.28GNGVDD343 pKa = 5.02TYY345 pKa = 11.67DD346 pKa = 3.56LSNYY350 pKa = 8.51ATDD353 pKa = 4.73LSIDD357 pKa = 3.98LAPGAFSTFSAAQLVNHH374 pKa = 7.11RR375 pKa = 11.84AYY377 pKa = 10.89SGGVAPAVGNVANALLYY394 pKa = 11.32NNDD397 pKa = 3.06TRR399 pKa = 11.84SLIEE403 pKa = 3.94NATGGSGNDD412 pKa = 3.6SIVGNQANNTLRR424 pKa = 11.84GNDD427 pKa = 3.68GGDD430 pKa = 3.37TLVGSFGSDD439 pKa = 2.64KK440 pKa = 11.07LYY442 pKa = 11.1GGSAADD448 pKa = 3.54TLYY451 pKa = 11.39GDD453 pKa = 5.7GIPGIPPGVEE463 pKa = 3.62LGDD466 pKa = 3.72GVVVKK471 pKa = 9.53PAGAGNISLATAIDD485 pKa = 3.7VTKK488 pKa = 10.49EE489 pKa = 3.39FSLASNANIIDD500 pKa = 3.98STATPHH506 pKa = 4.98VTISGTGDD514 pKa = 3.26GNRR517 pKa = 11.84DD518 pKa = 3.5YY519 pKa = 11.89YY520 pKa = 11.38KK521 pKa = 10.15LTVNAGTTLTFDD533 pKa = 3.54IDD535 pKa = 3.66NTSGVDD541 pKa = 3.49SFLFLYY547 pKa = 10.51DD548 pKa = 3.55PTSTAVAFDD557 pKa = 4.89DD558 pKa = 4.87DD559 pKa = 4.39SQVTEE564 pKa = 4.21GAGGSTSSFDD574 pKa = 3.85SYY576 pKa = 10.88LTYY579 pKa = 10.18QVKK582 pKa = 7.91TAGTYY587 pKa = 9.1YY588 pKa = 9.98IEE590 pKa = 4.12VGAYY594 pKa = 9.6VSSTSNGPIGSGALYY609 pKa = 10.23DD610 pKa = 4.02LQISAGGTPGSGGGGTGNDD629 pKa = 3.74TLDD632 pKa = 3.99GGTGDD637 pKa = 5.02DD638 pKa = 4.32SMAGGAGSDD647 pKa = 3.82VYY649 pKa = 10.94FVDD652 pKa = 3.59SAADD656 pKa = 3.6VVTEE660 pKa = 3.97TAGNGTADD668 pKa = 3.28RR669 pKa = 11.84VAARR673 pKa = 11.84ASYY676 pKa = 10.89ALAAGVSIEE685 pKa = 4.45LLTTTSSAGTTAIDD699 pKa = 4.26LSGNALKK706 pKa = 8.77QTTTGNAGANTLKK719 pKa = 10.95DD720 pKa = 3.47GGGVADD726 pKa = 4.77ALNGLGGDD734 pKa = 4.64DD735 pKa = 3.7IYY737 pKa = 11.58LVYY740 pKa = 10.65AAGTTVAEE748 pKa = 4.37AAGQGSDD755 pKa = 3.12RR756 pKa = 11.84VAAAVDD762 pKa = 4.22FALGAGVHH770 pKa = 6.02IEE772 pKa = 4.04VLTTTSSGGTAAIDD786 pKa = 3.53LTGNALQQDD795 pKa = 3.64ITGNAGDD802 pKa = 4.93NILHH806 pKa = 7.22DD807 pKa = 4.72GGAGAADD814 pKa = 3.82TLKK817 pKa = 11.17GLGGNDD823 pKa = 3.31TYY825 pKa = 11.04RR826 pKa = 11.84VYY828 pKa = 11.02NAADD832 pKa = 3.57VVVEE836 pKa = 4.33TAAQGVYY843 pKa = 10.11DD844 pKa = 3.98VVMAAVDD851 pKa = 3.8YY852 pKa = 10.97VLTAGAHH859 pKa = 4.62VEE861 pKa = 3.97RR862 pKa = 11.84LATNGTAGTSGIDD875 pKa = 3.3LTGNEE880 pKa = 4.09IGQEE884 pKa = 4.14VVGNSGANKK893 pKa = 10.1LDD895 pKa = 3.62GKK897 pKa = 11.17GGDD900 pKa = 3.74DD901 pKa = 3.59TMKK904 pKa = 11.04GLAGKK909 pKa = 8.04DD910 pKa = 3.35TLTGGTGKK918 pKa = 10.36DD919 pKa = 2.99LFVFSTALSASTNVDD934 pKa = 2.94TVTDD938 pKa = 3.94FNVPNDD944 pKa = 4.15TIGLDD949 pKa = 3.69DD950 pKa = 6.12AIFAALSLGTLVASAFWASTTGLAHH975 pKa = 7.11DD976 pKa = 4.06ASDD979 pKa = 4.26RR980 pKa = 11.84ILYY983 pKa = 7.57EE984 pKa = 3.95TDD986 pKa = 2.82TGALFYY992 pKa = 11.14DD993 pKa = 4.17ADD995 pKa = 3.97GTGAGARR1002 pKa = 11.84IQFAVLTGLPALTNADD1018 pKa = 3.56FAVII1022 pKa = 3.94
MM1 pKa = 8.34CMMCQTVSRR10 pKa = 11.84FASEE14 pKa = 4.83EE15 pKa = 3.39IDD17 pKa = 3.51APMPSGSVSVAPPQFDD33 pKa = 3.58QMRR36 pKa = 11.84PPYY39 pKa = 10.63DD40 pKa = 3.27MRR42 pKa = 11.84EE43 pKa = 4.01QEE45 pKa = 4.38PGDD48 pKa = 3.55AYY50 pKa = 10.77GAGRR54 pKa = 11.84TALISQVFDD63 pKa = 3.65AADD66 pKa = 3.21GRR68 pKa = 11.84YY69 pKa = 9.17EE70 pKa = 3.81FSGNQDD76 pKa = 1.95IDD78 pKa = 3.3ATLIGSRR85 pKa = 11.84WTLSNLTFSFPTSGTFYY102 pKa = 11.06ADD104 pKa = 2.81EE105 pKa = 5.3GYY107 pKa = 9.37PAGTEE112 pKa = 4.04PAEE115 pKa = 3.9HH116 pKa = 6.07VAFNAMQQDD125 pKa = 3.7AVRR128 pKa = 11.84YY129 pKa = 8.95ALSLVQSYY137 pKa = 9.71TNLTFTEE144 pKa = 4.53VAEE147 pKa = 4.47SATTHH152 pKa = 6.11AEE154 pKa = 3.8LRR156 pKa = 11.84FSQTSYY162 pKa = 11.71DD163 pKa = 4.2DD164 pKa = 3.8VPSAYY169 pKa = 10.38ANFPSSSEE177 pKa = 3.79QAGDD181 pKa = 3.08VWFGMTGQPFYY192 pKa = 10.0DD193 pKa = 4.08TPAMGNWGMATILHH207 pKa = 6.74EE208 pKa = 4.78IGHH211 pKa = 6.45ALGLKK216 pKa = 9.85HH217 pKa = 6.41GHH219 pKa = 5.78QDD221 pKa = 3.35YY222 pKa = 8.54TAVDD226 pKa = 3.99LAGEE230 pKa = 5.19GYY232 pKa = 10.68LDD234 pKa = 3.99PPGGGGPRR242 pKa = 11.84YY243 pKa = 10.14GSAEE247 pKa = 3.92LPANHH252 pKa = 7.57DD253 pKa = 3.76GQDD256 pKa = 3.41WSLMTYY262 pKa = 10.25RR263 pKa = 11.84SDD265 pKa = 3.25PGAPVAFQGEE275 pKa = 5.11GFNQPQTYY283 pKa = 7.44MQNDD287 pKa = 3.39IAALQYY293 pKa = 11.06LYY295 pKa = 11.07GANFDD300 pKa = 4.42TNSSATVYY308 pKa = 10.17SWNPTTGEE316 pKa = 3.81MSIDD320 pKa = 3.58GVGQGAPTSNKK331 pKa = 9.63ISMTLWDD338 pKa = 4.28GNGVDD343 pKa = 5.02TYY345 pKa = 11.67DD346 pKa = 3.56LSNYY350 pKa = 8.51ATDD353 pKa = 4.73LSIDD357 pKa = 3.98LAPGAFSTFSAAQLVNHH374 pKa = 7.11RR375 pKa = 11.84AYY377 pKa = 10.89SGGVAPAVGNVANALLYY394 pKa = 11.32NNDD397 pKa = 3.06TRR399 pKa = 11.84SLIEE403 pKa = 3.94NATGGSGNDD412 pKa = 3.6SIVGNQANNTLRR424 pKa = 11.84GNDD427 pKa = 3.68GGDD430 pKa = 3.37TLVGSFGSDD439 pKa = 2.64KK440 pKa = 11.07LYY442 pKa = 11.1GGSAADD448 pKa = 3.54TLYY451 pKa = 11.39GDD453 pKa = 5.7GIPGIPPGVEE463 pKa = 3.62LGDD466 pKa = 3.72GVVVKK471 pKa = 9.53PAGAGNISLATAIDD485 pKa = 3.7VTKK488 pKa = 10.49EE489 pKa = 3.39FSLASNANIIDD500 pKa = 3.98STATPHH506 pKa = 4.98VTISGTGDD514 pKa = 3.26GNRR517 pKa = 11.84DD518 pKa = 3.5YY519 pKa = 11.89YY520 pKa = 11.38KK521 pKa = 10.15LTVNAGTTLTFDD533 pKa = 3.54IDD535 pKa = 3.66NTSGVDD541 pKa = 3.49SFLFLYY547 pKa = 10.51DD548 pKa = 3.55PTSTAVAFDD557 pKa = 4.89DD558 pKa = 4.87DD559 pKa = 4.39SQVTEE564 pKa = 4.21GAGGSTSSFDD574 pKa = 3.85SYY576 pKa = 10.88LTYY579 pKa = 10.18QVKK582 pKa = 7.91TAGTYY587 pKa = 9.1YY588 pKa = 9.98IEE590 pKa = 4.12VGAYY594 pKa = 9.6VSSTSNGPIGSGALYY609 pKa = 10.23DD610 pKa = 4.02LQISAGGTPGSGGGGTGNDD629 pKa = 3.74TLDD632 pKa = 3.99GGTGDD637 pKa = 5.02DD638 pKa = 4.32SMAGGAGSDD647 pKa = 3.82VYY649 pKa = 10.94FVDD652 pKa = 3.59SAADD656 pKa = 3.6VVTEE660 pKa = 3.97TAGNGTADD668 pKa = 3.28RR669 pKa = 11.84VAARR673 pKa = 11.84ASYY676 pKa = 10.89ALAAGVSIEE685 pKa = 4.45LLTTTSSAGTTAIDD699 pKa = 4.26LSGNALKK706 pKa = 8.77QTTTGNAGANTLKK719 pKa = 10.95DD720 pKa = 3.47GGGVADD726 pKa = 4.77ALNGLGGDD734 pKa = 4.64DD735 pKa = 3.7IYY737 pKa = 11.58LVYY740 pKa = 10.65AAGTTVAEE748 pKa = 4.37AAGQGSDD755 pKa = 3.12RR756 pKa = 11.84VAAAVDD762 pKa = 4.22FALGAGVHH770 pKa = 6.02IEE772 pKa = 4.04VLTTTSSGGTAAIDD786 pKa = 3.53LTGNALQQDD795 pKa = 3.64ITGNAGDD802 pKa = 4.93NILHH806 pKa = 7.22DD807 pKa = 4.72GGAGAADD814 pKa = 3.82TLKK817 pKa = 11.17GLGGNDD823 pKa = 3.31TYY825 pKa = 11.04RR826 pKa = 11.84VYY828 pKa = 11.02NAADD832 pKa = 3.57VVVEE836 pKa = 4.33TAAQGVYY843 pKa = 10.11DD844 pKa = 3.98VVMAAVDD851 pKa = 3.8YY852 pKa = 10.97VLTAGAHH859 pKa = 4.62VEE861 pKa = 3.97RR862 pKa = 11.84LATNGTAGTSGIDD875 pKa = 3.3LTGNEE880 pKa = 4.09IGQEE884 pKa = 4.14VVGNSGANKK893 pKa = 10.1LDD895 pKa = 3.62GKK897 pKa = 11.17GGDD900 pKa = 3.74DD901 pKa = 3.59TMKK904 pKa = 11.04GLAGKK909 pKa = 8.04DD910 pKa = 3.35TLTGGTGKK918 pKa = 10.36DD919 pKa = 2.99LFVFSTALSASTNVDD934 pKa = 2.94TVTDD938 pKa = 3.94FNVPNDD944 pKa = 4.15TIGLDD949 pKa = 3.69DD950 pKa = 6.12AIFAALSLGTLVASAFWASTTGLAHH975 pKa = 7.11DD976 pKa = 4.06ASDD979 pKa = 4.26RR980 pKa = 11.84ILYY983 pKa = 7.57EE984 pKa = 3.95TDD986 pKa = 2.82TGALFYY992 pKa = 11.14DD993 pKa = 4.17ADD995 pKa = 3.97GTGAGARR1002 pKa = 11.84IQFAVLTGLPALTNADD1018 pKa = 3.56FAVII1022 pKa = 3.94
Molecular weight: 104.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P7S8P3|A0A2P7S8P3_9RHIZ NAD(P)-dependent oxidoreductase OS=Mesorhizobium ephedrae OX=2116701 GN=C7I84_14455 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84GVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84GVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1760207 |
29 |
3292 |
310.1 |
33.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.982 ± 0.05 | 0.781 ± 0.01 |
5.777 ± 0.03 | 5.743 ± 0.033 |
3.87 ± 0.023 | 9.027 ± 0.043 |
1.962 ± 0.017 | 5.116 ± 0.024 |
3.237 ± 0.03 | 9.888 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.449 ± 0.016 | 2.465 ± 0.02 |
5.164 ± 0.029 | 2.801 ± 0.015 |
7.272 ± 0.037 | 5.162 ± 0.025 |
5.107 ± 0.027 | 7.661 ± 0.025 |
1.323 ± 0.012 | 2.213 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
