
Apis mellifera associated microvirus 2
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
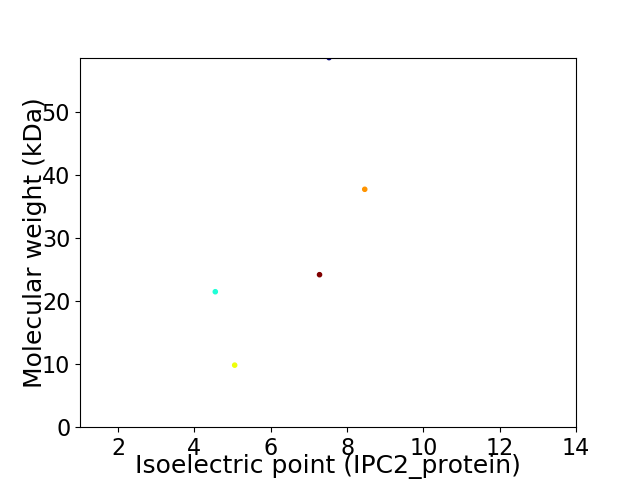
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
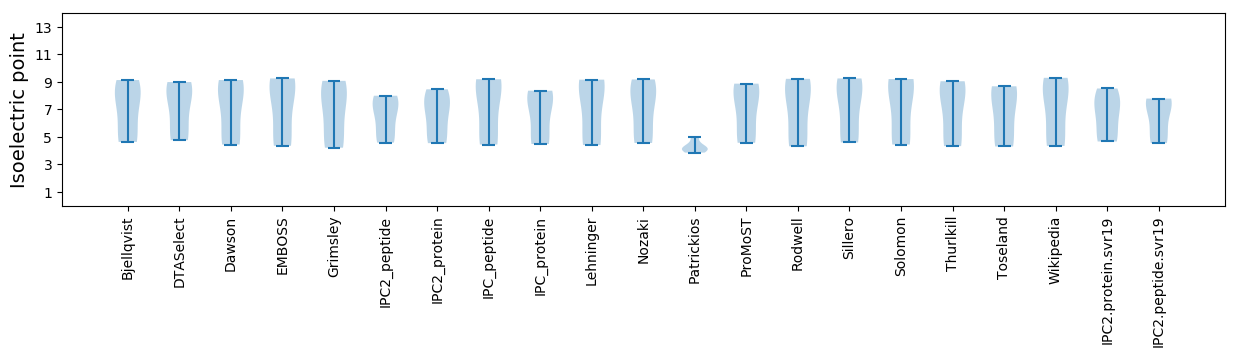
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTI6|A0A3S8UTI6_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 2 OX=2494747 PE=4 SV=1
MM1 pKa = 7.39AKK3 pKa = 10.16EE4 pKa = 4.44SVFLRR9 pKa = 11.84TPYY12 pKa = 10.64NYY14 pKa = 10.79DD15 pKa = 2.96RR16 pKa = 11.84MAVSDD21 pKa = 4.04EE22 pKa = 4.3TGLFCDD28 pKa = 5.37DD29 pKa = 4.4PSLTQQHH36 pKa = 6.69DD37 pKa = 3.6MADD40 pKa = 3.58CDD42 pKa = 3.48INNIVEE48 pKa = 4.06RR49 pKa = 11.84AARR52 pKa = 11.84TGFIPNAVGSPFVGDD67 pKa = 3.61LAINATDD74 pKa = 3.44FHH76 pKa = 6.54EE77 pKa = 4.41AMNIVVSARR86 pKa = 11.84QAFSEE91 pKa = 4.2LDD93 pKa = 3.09AKK95 pKa = 10.02TRR97 pKa = 11.84ARR99 pKa = 11.84FGNDD103 pKa = 2.88PGQLLEE109 pKa = 4.08FLQDD113 pKa = 3.05EE114 pKa = 4.69GNYY117 pKa = 10.81DD118 pKa = 3.73EE119 pKa = 5.32ASKK122 pKa = 11.03LGLVPPRR129 pKa = 11.84EE130 pKa = 3.96EE131 pKa = 3.87LSIGEE136 pKa = 4.0RR137 pKa = 11.84SANKK141 pKa = 9.58SSRR144 pKa = 11.84RR145 pKa = 11.84SSPVEE150 pKa = 3.6NSASSDD156 pKa = 3.45FDD158 pKa = 4.88SDD160 pKa = 4.08DD161 pKa = 3.57SQDD164 pKa = 3.83RR165 pKa = 11.84VAEE168 pKa = 4.13LQAQLQRR175 pKa = 11.84LQGSSSKK182 pKa = 10.62KK183 pKa = 9.41GVSSPFRR190 pKa = 11.84RR191 pKa = 11.84GSGDD195 pKa = 3.26EE196 pKa = 3.89
MM1 pKa = 7.39AKK3 pKa = 10.16EE4 pKa = 4.44SVFLRR9 pKa = 11.84TPYY12 pKa = 10.64NYY14 pKa = 10.79DD15 pKa = 2.96RR16 pKa = 11.84MAVSDD21 pKa = 4.04EE22 pKa = 4.3TGLFCDD28 pKa = 5.37DD29 pKa = 4.4PSLTQQHH36 pKa = 6.69DD37 pKa = 3.6MADD40 pKa = 3.58CDD42 pKa = 3.48INNIVEE48 pKa = 4.06RR49 pKa = 11.84AARR52 pKa = 11.84TGFIPNAVGSPFVGDD67 pKa = 3.61LAINATDD74 pKa = 3.44FHH76 pKa = 6.54EE77 pKa = 4.41AMNIVVSARR86 pKa = 11.84QAFSEE91 pKa = 4.2LDD93 pKa = 3.09AKK95 pKa = 10.02TRR97 pKa = 11.84ARR99 pKa = 11.84FGNDD103 pKa = 2.88PGQLLEE109 pKa = 4.08FLQDD113 pKa = 3.05EE114 pKa = 4.69GNYY117 pKa = 10.81DD118 pKa = 3.73EE119 pKa = 5.32ASKK122 pKa = 11.03LGLVPPRR129 pKa = 11.84EE130 pKa = 3.96EE131 pKa = 3.87LSIGEE136 pKa = 4.0RR137 pKa = 11.84SANKK141 pKa = 9.58SSRR144 pKa = 11.84RR145 pKa = 11.84SSPVEE150 pKa = 3.6NSASSDD156 pKa = 3.45FDD158 pKa = 4.88SDD160 pKa = 4.08DD161 pKa = 3.57SQDD164 pKa = 3.83RR165 pKa = 11.84VAEE168 pKa = 4.13LQAQLQRR175 pKa = 11.84LQGSSSKK182 pKa = 10.62KK183 pKa = 9.41GVSSPFRR190 pKa = 11.84RR191 pKa = 11.84GSGDD195 pKa = 3.26EE196 pKa = 3.89
Molecular weight: 21.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTJ6|A0A3S8UTJ6_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 2 OX=2494747 PE=3 SV=1
MM1 pKa = 7.08VCYY4 pKa = 9.87FPITGYY10 pKa = 10.3QSQPGAPLVFGGIAVNADD28 pKa = 3.2KK29 pKa = 10.99TPIKK33 pKa = 10.17VPCGRR38 pKa = 11.84CEE40 pKa = 4.19GCNEE44 pKa = 3.9RR45 pKa = 11.84YY46 pKa = 10.14ARR48 pKa = 11.84DD49 pKa = 2.92WAIRR53 pKa = 11.84MMHH56 pKa = 6.23EE57 pKa = 3.72ASLYY61 pKa = 9.82KK62 pKa = 10.33HH63 pKa = 6.17NCFITLTYY71 pKa = 10.87NNEE74 pKa = 3.93NLPPDD79 pKa = 3.59RR80 pKa = 11.84SLHH83 pKa = 5.35YY84 pKa = 10.49EE85 pKa = 3.99HH86 pKa = 6.72FQEE89 pKa = 4.15FMKK92 pKa = 10.52RR93 pKa = 11.84FRR95 pKa = 11.84EE96 pKa = 4.1AHH98 pKa = 5.53QGCDD102 pKa = 3.75LVSHH106 pKa = 7.22PYY108 pKa = 9.65FGKK111 pKa = 9.83IDD113 pKa = 3.63KK114 pKa = 8.96KK115 pKa = 9.21TGKK118 pKa = 9.25PYY120 pKa = 10.39PEE122 pKa = 4.82FYY124 pKa = 10.48RR125 pKa = 11.84PIRR128 pKa = 11.84HH129 pKa = 5.26YY130 pKa = 11.04VAGEE134 pKa = 3.77YY135 pKa = 10.25GGKK138 pKa = 9.88LGRR141 pKa = 11.84PHH143 pKa = 5.98WHH145 pKa = 5.94ACIFNFDD152 pKa = 3.78FADD155 pKa = 3.69KK156 pKa = 10.53YY157 pKa = 10.49IWEE160 pKa = 4.47TSTSGEE166 pKa = 3.92PLYY169 pKa = 10.52RR170 pKa = 11.84SPSLEE175 pKa = 3.84QLWPFGYY182 pKa = 10.38SSIGSVTFQSAAYY195 pKa = 8.01VARR198 pKa = 11.84YY199 pKa = 9.05INKK202 pKa = 9.28KK203 pKa = 9.91VKK205 pKa = 10.17GNQADD210 pKa = 3.83DD211 pKa = 3.73HH212 pKa = 6.28YY213 pKa = 11.06AWHH216 pKa = 7.13DD217 pKa = 3.7PEE219 pKa = 4.4TGEE222 pKa = 4.38VFWRR226 pKa = 11.84RR227 pKa = 11.84PEE229 pKa = 3.84FSKK232 pKa = 10.68MSLKK236 pKa = 10.58PGIGAGWLAKK246 pKa = 10.52YY247 pKa = 8.65KK248 pKa = 10.41TDD250 pKa = 4.13VFPHH254 pKa = 6.02DD255 pKa = 5.21HH256 pKa = 6.99IISDD260 pKa = 4.07GRR262 pKa = 11.84PLPVPRR268 pKa = 11.84YY269 pKa = 8.34YY270 pKa = 11.19SKK272 pKa = 10.48IYY274 pKa = 10.21QCTNPLEE281 pKa = 4.32WDD283 pKa = 3.44AVAHH287 pKa = 5.1EE288 pKa = 5.1RR289 pKa = 11.84YY290 pKa = 9.75VKK292 pKa = 10.4SRR294 pKa = 11.84QTLDD298 pKa = 3.61DD299 pKa = 3.78NTPEE303 pKa = 3.8RR304 pKa = 11.84LAVKK308 pKa = 10.13RR309 pKa = 11.84QVHH312 pKa = 5.43LAKK315 pKa = 10.67LSRR318 pKa = 11.84LKK320 pKa = 10.17RR321 pKa = 11.84TLKK324 pKa = 10.74
MM1 pKa = 7.08VCYY4 pKa = 9.87FPITGYY10 pKa = 10.3QSQPGAPLVFGGIAVNADD28 pKa = 3.2KK29 pKa = 10.99TPIKK33 pKa = 10.17VPCGRR38 pKa = 11.84CEE40 pKa = 4.19GCNEE44 pKa = 3.9RR45 pKa = 11.84YY46 pKa = 10.14ARR48 pKa = 11.84DD49 pKa = 2.92WAIRR53 pKa = 11.84MMHH56 pKa = 6.23EE57 pKa = 3.72ASLYY61 pKa = 9.82KK62 pKa = 10.33HH63 pKa = 6.17NCFITLTYY71 pKa = 10.87NNEE74 pKa = 3.93NLPPDD79 pKa = 3.59RR80 pKa = 11.84SLHH83 pKa = 5.35YY84 pKa = 10.49EE85 pKa = 3.99HH86 pKa = 6.72FQEE89 pKa = 4.15FMKK92 pKa = 10.52RR93 pKa = 11.84FRR95 pKa = 11.84EE96 pKa = 4.1AHH98 pKa = 5.53QGCDD102 pKa = 3.75LVSHH106 pKa = 7.22PYY108 pKa = 9.65FGKK111 pKa = 9.83IDD113 pKa = 3.63KK114 pKa = 8.96KK115 pKa = 9.21TGKK118 pKa = 9.25PYY120 pKa = 10.39PEE122 pKa = 4.82FYY124 pKa = 10.48RR125 pKa = 11.84PIRR128 pKa = 11.84HH129 pKa = 5.26YY130 pKa = 11.04VAGEE134 pKa = 3.77YY135 pKa = 10.25GGKK138 pKa = 9.88LGRR141 pKa = 11.84PHH143 pKa = 5.98WHH145 pKa = 5.94ACIFNFDD152 pKa = 3.78FADD155 pKa = 3.69KK156 pKa = 10.53YY157 pKa = 10.49IWEE160 pKa = 4.47TSTSGEE166 pKa = 3.92PLYY169 pKa = 10.52RR170 pKa = 11.84SPSLEE175 pKa = 3.84QLWPFGYY182 pKa = 10.38SSIGSVTFQSAAYY195 pKa = 8.01VARR198 pKa = 11.84YY199 pKa = 9.05INKK202 pKa = 9.28KK203 pKa = 9.91VKK205 pKa = 10.17GNQADD210 pKa = 3.83DD211 pKa = 3.73HH212 pKa = 6.28YY213 pKa = 11.06AWHH216 pKa = 7.13DD217 pKa = 3.7PEE219 pKa = 4.4TGEE222 pKa = 4.38VFWRR226 pKa = 11.84RR227 pKa = 11.84PEE229 pKa = 3.84FSKK232 pKa = 10.68MSLKK236 pKa = 10.58PGIGAGWLAKK246 pKa = 10.52YY247 pKa = 8.65KK248 pKa = 10.41TDD250 pKa = 4.13VFPHH254 pKa = 6.02DD255 pKa = 5.21HH256 pKa = 6.99IISDD260 pKa = 4.07GRR262 pKa = 11.84PLPVPRR268 pKa = 11.84YY269 pKa = 8.34YY270 pKa = 11.19SKK272 pKa = 10.48IYY274 pKa = 10.21QCTNPLEE281 pKa = 4.32WDD283 pKa = 3.44AVAHH287 pKa = 5.1EE288 pKa = 5.1RR289 pKa = 11.84YY290 pKa = 9.75VKK292 pKa = 10.4SRR294 pKa = 11.84QTLDD298 pKa = 3.61DD299 pKa = 3.78NTPEE303 pKa = 3.8RR304 pKa = 11.84LAVKK308 pKa = 10.13RR309 pKa = 11.84QVHH312 pKa = 5.43LAKK315 pKa = 10.67LSRR318 pKa = 11.84LKK320 pKa = 10.17RR321 pKa = 11.84TLKK324 pKa = 10.74
Molecular weight: 37.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1366 |
86 |
533 |
273.2 |
30.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.81 ± 1.68 | 1.025 ± 0.393 |
5.93 ± 1.155 | 4.173 ± 0.981 |
5.198 ± 0.422 | 7.101 ± 0.779 |
2.709 ± 0.581 | 4.026 ± 0.292 |
4.246 ± 1.058 | 7.394 ± 0.409 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.343 ± 0.477 | 4.832 ± 0.576 |
5.49 ± 0.681 | 4.612 ± 0.538 |
5.637 ± 0.703 | 7.687 ± 0.986 |
6.735 ± 1.291 | 5.564 ± 0.49 |
1.464 ± 0.376 | 4.026 ± 1.072 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
