
Shuangao sobemo-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.57
Get precalculated fractions of proteins
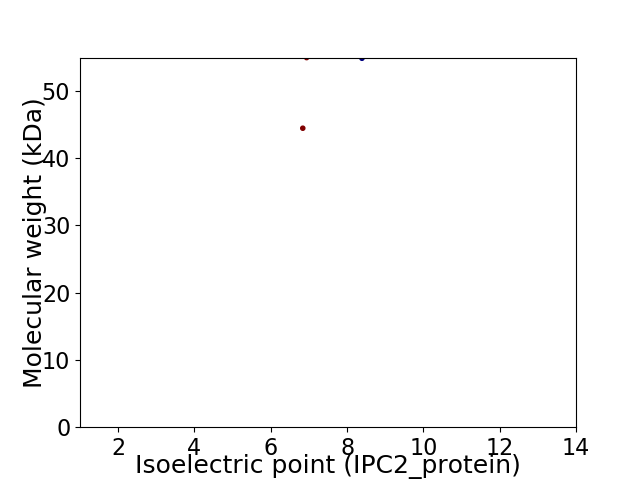
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
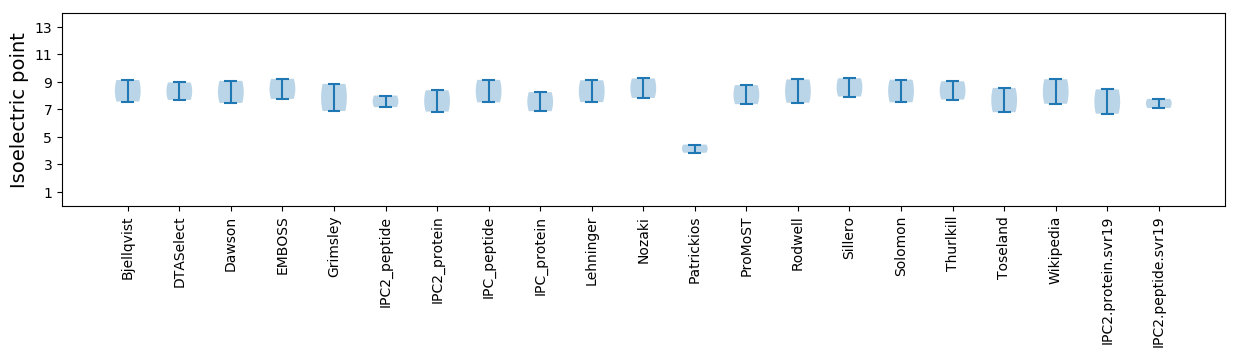
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEH8|A0A1L3KEH8_9VIRU RNA-directed RNA polymerase OS=Shuangao sobemo-like virus 3 OX=1923476 PE=4 SV=1
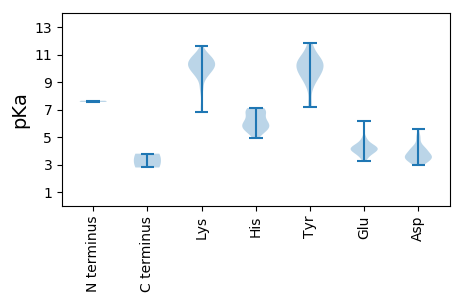
MM1 pKa = 7.55EE2 pKa = 6.14EE3 pKa = 3.96IYY5 pKa = 11.07SPVVWNLPDD14 pKa = 3.92NYY16 pKa = 10.51LSYY19 pKa = 11.57DD20 pKa = 3.66SFVKK24 pKa = 10.21VCYY27 pKa = 10.4GLDD30 pKa = 3.56FRR32 pKa = 11.84SSPGVPYY39 pKa = 10.38CYY41 pKa = 10.02DD42 pKa = 3.6YY43 pKa = 9.32PTVGDD48 pKa = 3.61WFHH51 pKa = 7.11RR52 pKa = 11.84KK53 pKa = 9.36ALDD56 pKa = 3.51LSEE59 pKa = 4.39QRR61 pKa = 11.84LQILWLDD68 pKa = 3.69VQDD71 pKa = 5.57LINGKK76 pKa = 9.47LDD78 pKa = 4.44PILRR82 pKa = 11.84YY83 pKa = 9.27FIKK86 pKa = 10.58RR87 pKa = 11.84EE88 pKa = 3.62PHH90 pKa = 5.42KK91 pKa = 10.54KK92 pKa = 10.1KK93 pKa = 10.7KK94 pKa = 9.48IAQKK98 pKa = 9.52RR99 pKa = 11.84WRR101 pKa = 11.84LIASTPLNVQVLWHH115 pKa = 6.09MLFDD119 pKa = 4.09FQNDD123 pKa = 3.43KK124 pKa = 10.91EE125 pKa = 4.24IEE127 pKa = 4.14QVYY130 pKa = 10.14SIPSKK135 pKa = 10.31QGMSLVHH142 pKa = 5.8GQWKK146 pKa = 9.58FYY148 pKa = 9.77KK149 pKa = 10.15RR150 pKa = 11.84QWKK153 pKa = 9.86SKK155 pKa = 9.69GFNFCVDD162 pKa = 3.39AEE164 pKa = 4.26AWDD167 pKa = 3.36WTMPYY172 pKa = 10.25WLILWDD178 pKa = 3.57LDD180 pKa = 3.09FRR182 pKa = 11.84YY183 pKa = 10.65RR184 pKa = 11.84MGRR187 pKa = 11.84GRR189 pKa = 11.84KK190 pKa = 7.26MDD192 pKa = 3.42EE193 pKa = 3.58WRR195 pKa = 11.84LLALNLIQYY204 pKa = 7.67MFEE207 pKa = 4.32KK208 pKa = 10.64PLVCLSDD215 pKa = 3.41GSLYY219 pKa = 10.48RR220 pKa = 11.84QLLAGIMKK228 pKa = 9.72SGCVNTISTNSHH240 pKa = 4.93GQIMDD245 pKa = 4.55HH246 pKa = 6.89IIVCVRR252 pKa = 11.84EE253 pKa = 4.04NLPIYY258 pKa = 8.62PLPDD262 pKa = 3.3ACGDD266 pKa = 3.46DD267 pKa = 4.07KK268 pKa = 11.61YY269 pKa = 11.42QNEE272 pKa = 4.14INAGSVEE279 pKa = 4.33AFSKK283 pKa = 10.46IGVKK287 pKa = 9.85IKK289 pKa = 10.79LMEE292 pKa = 5.02KK293 pKa = 8.44GHH295 pKa = 6.68DD296 pKa = 4.21FIGHH300 pKa = 5.49DD301 pKa = 4.03HH302 pKa = 6.47SLEE305 pKa = 4.34SGPVPLYY312 pKa = 10.31FDD314 pKa = 3.22KK315 pKa = 11.18HH316 pKa = 5.74LFSYY320 pKa = 10.62CIEE323 pKa = 4.15KK324 pKa = 10.79PEE326 pKa = 3.9FLSEE330 pKa = 4.17FLEE333 pKa = 4.67SMCFLYY339 pKa = 9.74THH341 pKa = 5.95SEE343 pKa = 4.19KK344 pKa = 10.8FSFWYY349 pKa = 10.27DD350 pKa = 3.08LALKK354 pKa = 10.27HH355 pKa = 5.87NSRR358 pKa = 11.84VKK360 pKa = 10.14SRR362 pKa = 11.84FYY364 pKa = 10.62YY365 pKa = 10.41KK366 pKa = 10.73YY367 pKa = 9.74MYY369 pKa = 9.36DD370 pKa = 3.25IPVAA374 pKa = 3.79
MM1 pKa = 7.55EE2 pKa = 6.14EE3 pKa = 3.96IYY5 pKa = 11.07SPVVWNLPDD14 pKa = 3.92NYY16 pKa = 10.51LSYY19 pKa = 11.57DD20 pKa = 3.66SFVKK24 pKa = 10.21VCYY27 pKa = 10.4GLDD30 pKa = 3.56FRR32 pKa = 11.84SSPGVPYY39 pKa = 10.38CYY41 pKa = 10.02DD42 pKa = 3.6YY43 pKa = 9.32PTVGDD48 pKa = 3.61WFHH51 pKa = 7.11RR52 pKa = 11.84KK53 pKa = 9.36ALDD56 pKa = 3.51LSEE59 pKa = 4.39QRR61 pKa = 11.84LQILWLDD68 pKa = 3.69VQDD71 pKa = 5.57LINGKK76 pKa = 9.47LDD78 pKa = 4.44PILRR82 pKa = 11.84YY83 pKa = 9.27FIKK86 pKa = 10.58RR87 pKa = 11.84EE88 pKa = 3.62PHH90 pKa = 5.42KK91 pKa = 10.54KK92 pKa = 10.1KK93 pKa = 10.7KK94 pKa = 9.48IAQKK98 pKa = 9.52RR99 pKa = 11.84WRR101 pKa = 11.84LIASTPLNVQVLWHH115 pKa = 6.09MLFDD119 pKa = 4.09FQNDD123 pKa = 3.43KK124 pKa = 10.91EE125 pKa = 4.24IEE127 pKa = 4.14QVYY130 pKa = 10.14SIPSKK135 pKa = 10.31QGMSLVHH142 pKa = 5.8GQWKK146 pKa = 9.58FYY148 pKa = 9.77KK149 pKa = 10.15RR150 pKa = 11.84QWKK153 pKa = 9.86SKK155 pKa = 9.69GFNFCVDD162 pKa = 3.39AEE164 pKa = 4.26AWDD167 pKa = 3.36WTMPYY172 pKa = 10.25WLILWDD178 pKa = 3.57LDD180 pKa = 3.09FRR182 pKa = 11.84YY183 pKa = 10.65RR184 pKa = 11.84MGRR187 pKa = 11.84GRR189 pKa = 11.84KK190 pKa = 7.26MDD192 pKa = 3.42EE193 pKa = 3.58WRR195 pKa = 11.84LLALNLIQYY204 pKa = 7.67MFEE207 pKa = 4.32KK208 pKa = 10.64PLVCLSDD215 pKa = 3.41GSLYY219 pKa = 10.48RR220 pKa = 11.84QLLAGIMKK228 pKa = 9.72SGCVNTISTNSHH240 pKa = 4.93GQIMDD245 pKa = 4.55HH246 pKa = 6.89IIVCVRR252 pKa = 11.84EE253 pKa = 4.04NLPIYY258 pKa = 8.62PLPDD262 pKa = 3.3ACGDD266 pKa = 3.46DD267 pKa = 4.07KK268 pKa = 11.61YY269 pKa = 11.42QNEE272 pKa = 4.14INAGSVEE279 pKa = 4.33AFSKK283 pKa = 10.46IGVKK287 pKa = 9.85IKK289 pKa = 10.79LMEE292 pKa = 5.02KK293 pKa = 8.44GHH295 pKa = 6.68DD296 pKa = 4.21FIGHH300 pKa = 5.49DD301 pKa = 4.03HH302 pKa = 6.47SLEE305 pKa = 4.34SGPVPLYY312 pKa = 10.31FDD314 pKa = 3.22KK315 pKa = 11.18HH316 pKa = 5.74LFSYY320 pKa = 10.62CIEE323 pKa = 4.15KK324 pKa = 10.79PEE326 pKa = 3.9FLSEE330 pKa = 4.17FLEE333 pKa = 4.67SMCFLYY339 pKa = 9.74THH341 pKa = 5.95SEE343 pKa = 4.19KK344 pKa = 10.8FSFWYY349 pKa = 10.27DD350 pKa = 3.08LALKK354 pKa = 10.27HH355 pKa = 5.87NSRR358 pKa = 11.84VKK360 pKa = 10.14SRR362 pKa = 11.84FYY364 pKa = 10.62YY365 pKa = 10.41KK366 pKa = 10.73YY367 pKa = 9.74MYY369 pKa = 9.36DD370 pKa = 3.25IPVAA374 pKa = 3.79
Molecular weight: 44.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEH8|A0A1L3KEH8_9VIRU RNA-directed RNA polymerase OS=Shuangao sobemo-like virus 3 OX=1923476 PE=4 SV=1
MM1 pKa = 7.64AFLSSVLSKK10 pKa = 10.83INWSTVVPVAFSIANTAAVAYY31 pKa = 7.17FACQGTTSVVVIPEE45 pKa = 4.33EE46 pKa = 4.1VPSWKK51 pKa = 9.97NRR53 pKa = 11.84FSLSSIIPYY62 pKa = 9.54FEE64 pKa = 3.76KK65 pKa = 10.05QVPIRR70 pKa = 11.84RR71 pKa = 11.84VAITCCGVYY80 pKa = 9.74VFWFCYY86 pKa = 8.82RR87 pKa = 11.84TGLFSKK93 pKa = 9.95FFSFLRR99 pKa = 11.84YY100 pKa = 7.24MAPGFNYY107 pKa = 9.72IKK109 pKa = 10.52FKK111 pKa = 10.43LTGHH115 pKa = 5.94PEE117 pKa = 3.27IRR119 pKa = 11.84IEE121 pKa = 4.01IDD123 pKa = 2.97PTEE126 pKa = 4.03TSTRR130 pKa = 11.84IIQEE134 pKa = 4.17SIRR137 pKa = 11.84AGSTEE142 pKa = 4.03SAANPPHH149 pKa = 5.71FQCMIGYY156 pKa = 9.01VDD158 pKa = 3.77SGVFTAVGAAIRR170 pKa = 11.84IHH172 pKa = 7.06DD173 pKa = 4.15VLVMPRR179 pKa = 11.84HH180 pKa = 6.23VYY182 pKa = 8.7DD183 pKa = 3.23QCMEE187 pKa = 4.01RR188 pKa = 11.84PAAYY192 pKa = 10.16GNRR195 pKa = 11.84TKK197 pKa = 11.07NIVNLVNFEE206 pKa = 4.63CIDD209 pKa = 3.64MDD211 pKa = 4.0TDD213 pKa = 5.0LIMVKK218 pKa = 6.85TTKK221 pKa = 10.43DD222 pKa = 3.08ALATIGMAIPSIQNCIPIAGTNVNIVGIEE251 pKa = 3.99NAGTTGNLQLSEE263 pKa = 4.52VNFGCTVYY271 pKa = 10.72HH272 pKa = 6.26GTTKK276 pKa = 10.34PGYY279 pKa = 9.93SGAAYY284 pKa = 9.53FAGPRR289 pKa = 11.84LVGMHH294 pKa = 5.47SHH296 pKa = 6.92GGPLNGGYY304 pKa = 9.46SASFILAMVNHH315 pKa = 6.84FLSKK319 pKa = 10.27IPEE322 pKa = 4.71DD323 pKa = 3.63SDD325 pKa = 3.03KK326 pKa = 11.09WLAQQFRR333 pKa = 11.84AGKK336 pKa = 9.36EE337 pKa = 3.67IKK339 pKa = 10.01FDD341 pKa = 4.25LGWGSLDD348 pKa = 3.47TEE350 pKa = 4.95RR351 pKa = 11.84IQIGGEE357 pKa = 3.31FHH359 pKa = 7.0IVQKK363 pKa = 10.27RR364 pKa = 11.84SCVSAIGKK372 pKa = 10.01DD373 pKa = 3.67YY374 pKa = 11.2GNHH377 pKa = 5.41VNDD380 pKa = 4.97DD381 pKa = 4.08GYY383 pKa = 11.83LKK385 pKa = 10.76FKK387 pKa = 10.92FDD389 pKa = 3.99GQYY392 pKa = 10.81QDD394 pKa = 5.07YY395 pKa = 10.8SDD397 pKa = 4.21EE398 pKa = 4.4DD399 pKa = 3.73SSRR402 pKa = 11.84GRR404 pKa = 11.84ARR406 pKa = 11.84SRR408 pKa = 11.84RR409 pKa = 11.84GRR411 pKa = 11.84KK412 pKa = 8.47IKK414 pKa = 10.35RR415 pKa = 11.84NRR417 pKa = 11.84FRR419 pKa = 11.84SEE421 pKa = 3.83SPINEE426 pKa = 3.82SAIPKK431 pKa = 9.17NEE433 pKa = 3.6KK434 pKa = 10.27SRR436 pKa = 11.84GSNTSGKK443 pKa = 9.92SEE445 pKa = 3.89ASADD449 pKa = 3.66QLINEE454 pKa = 4.5SLRR457 pKa = 11.84VEE459 pKa = 4.23LMNLISSAIRR469 pKa = 11.84SSMKK473 pKa = 8.56RR474 pKa = 11.84TAYY477 pKa = 10.06RR478 pKa = 11.84KK479 pKa = 10.0SPPTPNTNPSNSTPNTSS496 pKa = 2.78
MM1 pKa = 7.64AFLSSVLSKK10 pKa = 10.83INWSTVVPVAFSIANTAAVAYY31 pKa = 7.17FACQGTTSVVVIPEE45 pKa = 4.33EE46 pKa = 4.1VPSWKK51 pKa = 9.97NRR53 pKa = 11.84FSLSSIIPYY62 pKa = 9.54FEE64 pKa = 3.76KK65 pKa = 10.05QVPIRR70 pKa = 11.84RR71 pKa = 11.84VAITCCGVYY80 pKa = 9.74VFWFCYY86 pKa = 8.82RR87 pKa = 11.84TGLFSKK93 pKa = 9.95FFSFLRR99 pKa = 11.84YY100 pKa = 7.24MAPGFNYY107 pKa = 9.72IKK109 pKa = 10.52FKK111 pKa = 10.43LTGHH115 pKa = 5.94PEE117 pKa = 3.27IRR119 pKa = 11.84IEE121 pKa = 4.01IDD123 pKa = 2.97PTEE126 pKa = 4.03TSTRR130 pKa = 11.84IIQEE134 pKa = 4.17SIRR137 pKa = 11.84AGSTEE142 pKa = 4.03SAANPPHH149 pKa = 5.71FQCMIGYY156 pKa = 9.01VDD158 pKa = 3.77SGVFTAVGAAIRR170 pKa = 11.84IHH172 pKa = 7.06DD173 pKa = 4.15VLVMPRR179 pKa = 11.84HH180 pKa = 6.23VYY182 pKa = 8.7DD183 pKa = 3.23QCMEE187 pKa = 4.01RR188 pKa = 11.84PAAYY192 pKa = 10.16GNRR195 pKa = 11.84TKK197 pKa = 11.07NIVNLVNFEE206 pKa = 4.63CIDD209 pKa = 3.64MDD211 pKa = 4.0TDD213 pKa = 5.0LIMVKK218 pKa = 6.85TTKK221 pKa = 10.43DD222 pKa = 3.08ALATIGMAIPSIQNCIPIAGTNVNIVGIEE251 pKa = 3.99NAGTTGNLQLSEE263 pKa = 4.52VNFGCTVYY271 pKa = 10.72HH272 pKa = 6.26GTTKK276 pKa = 10.34PGYY279 pKa = 9.93SGAAYY284 pKa = 9.53FAGPRR289 pKa = 11.84LVGMHH294 pKa = 5.47SHH296 pKa = 6.92GGPLNGGYY304 pKa = 9.46SASFILAMVNHH315 pKa = 6.84FLSKK319 pKa = 10.27IPEE322 pKa = 4.71DD323 pKa = 3.63SDD325 pKa = 3.03KK326 pKa = 11.09WLAQQFRR333 pKa = 11.84AGKK336 pKa = 9.36EE337 pKa = 3.67IKK339 pKa = 10.01FDD341 pKa = 4.25LGWGSLDD348 pKa = 3.47TEE350 pKa = 4.95RR351 pKa = 11.84IQIGGEE357 pKa = 3.31FHH359 pKa = 7.0IVQKK363 pKa = 10.27RR364 pKa = 11.84SCVSAIGKK372 pKa = 10.01DD373 pKa = 3.67YY374 pKa = 11.2GNHH377 pKa = 5.41VNDD380 pKa = 4.97DD381 pKa = 4.08GYY383 pKa = 11.83LKK385 pKa = 10.76FKK387 pKa = 10.92FDD389 pKa = 3.99GQYY392 pKa = 10.81QDD394 pKa = 5.07YY395 pKa = 10.8SDD397 pKa = 4.21EE398 pKa = 4.4DD399 pKa = 3.73SSRR402 pKa = 11.84GRR404 pKa = 11.84ARR406 pKa = 11.84SRR408 pKa = 11.84RR409 pKa = 11.84GRR411 pKa = 11.84KK412 pKa = 8.47IKK414 pKa = 10.35RR415 pKa = 11.84NRR417 pKa = 11.84FRR419 pKa = 11.84SEE421 pKa = 3.83SPINEE426 pKa = 3.82SAIPKK431 pKa = 9.17NEE433 pKa = 3.6KK434 pKa = 10.27SRR436 pKa = 11.84GSNTSGKK443 pKa = 9.92SEE445 pKa = 3.89ASADD449 pKa = 3.66QLINEE454 pKa = 4.5SLRR457 pKa = 11.84VEE459 pKa = 4.23LMNLISSAIRR469 pKa = 11.84SSMKK473 pKa = 8.56RR474 pKa = 11.84TAYY477 pKa = 10.06RR478 pKa = 11.84KK479 pKa = 10.0SPPTPNTNPSNSTPNTSS496 pKa = 2.78
Molecular weight: 54.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
870 |
374 |
496 |
435.0 |
49.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.517 ± 1.434 | 2.184 ± 0.138 |
5.287 ± 1.034 | 4.828 ± 0.157 |
5.402 ± 0.132 | 6.552 ± 0.914 |
2.529 ± 0.422 | 7.241 ± 0.678 |
6.207 ± 0.961 | 7.241 ± 1.979 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.759 ± 0.279 | 4.828 ± 0.839 |
4.943 ± 0.081 | 3.218 ± 0.326 |
5.402 ± 0.532 | 8.736 ± 0.942 |
4.138 ± 1.574 | 6.092 ± 0.296 |
2.069 ± 0.874 | 4.828 ± 0.987 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
