
Kangiella koreensis (strain DSM 16069 / KCTC 12182 / SW-125)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Kangiellaceae; Kangiella; Kangiella koreensis
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
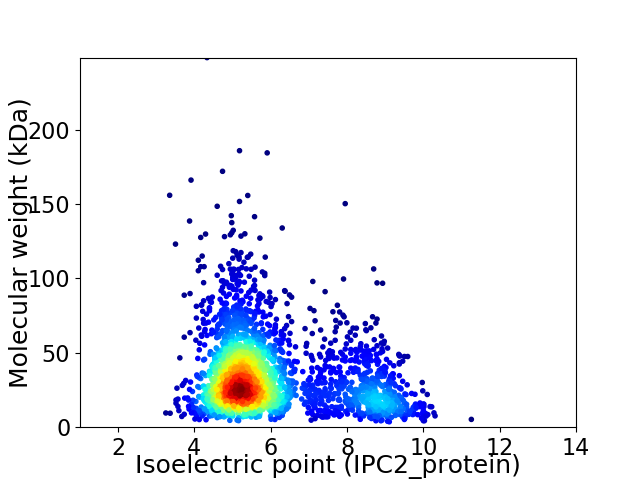
Virtual 2D-PAGE plot for 2630 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7RB18|C7RB18_KANKD Helicase c2 OS=Kangiella koreensis (strain DSM 16069 / KCTC 12182 / SW-125) OX=523791 GN=Kkor_1041 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.36KK3 pKa = 10.32VIPFLASALAFLPQSVISQDD23 pKa = 3.6TILVDD28 pKa = 3.12TGEE31 pKa = 4.59VYY33 pKa = 10.94GHH35 pKa = 6.17GPYY38 pKa = 10.5YY39 pKa = 10.62GVEE42 pKa = 4.13AYY44 pKa = 10.56GNASYY49 pKa = 11.76VNDD52 pKa = 3.92GSKK55 pKa = 10.15EE56 pKa = 3.79IKK58 pKa = 10.76VFDD61 pKa = 3.91VSDD64 pKa = 3.69ASNITLSGEE73 pKa = 4.28VEE75 pKa = 4.43VGCTVDD81 pKa = 6.3DD82 pKa = 4.88LDD84 pKa = 6.25LDD86 pKa = 4.2GATLVAKK93 pKa = 10.36CPFEE97 pKa = 3.55IHH99 pKa = 6.81FYY101 pKa = 11.4DD102 pKa = 4.66LTDD105 pKa = 3.43SLNPEE110 pKa = 4.2LVGSYY115 pKa = 10.82DD116 pKa = 3.52STSYY120 pKa = 11.18AVNSVEE126 pKa = 4.59LSGDD130 pKa = 3.08RR131 pKa = 11.84LYY133 pKa = 10.63IGGGNTDD140 pKa = 2.58IVIFNASDD148 pKa = 3.57LSNIQEE154 pKa = 4.47LNSQTFDD161 pKa = 3.64SISTGEE167 pKa = 4.21LKK169 pKa = 10.65KK170 pKa = 10.76SANFVYY176 pKa = 10.45SISDD180 pKa = 3.54FDD182 pKa = 3.91TVKK185 pKa = 10.59IFDD188 pKa = 3.67VSNEE192 pKa = 3.68EE193 pKa = 4.35NIIKK197 pKa = 9.71TSEE200 pKa = 3.79INATATLFKK209 pKa = 10.8DD210 pKa = 3.42VLIIDD215 pKa = 3.64NTLYY219 pKa = 10.41IAATDD224 pKa = 3.67GLRR227 pKa = 11.84VYY229 pKa = 10.63DD230 pKa = 4.12VTNMASPSYY239 pKa = 11.0VKK241 pKa = 10.08TIDD244 pKa = 3.17SGSFFNTIDD253 pKa = 4.02LLNALHH259 pKa = 6.45VVGDD263 pKa = 3.49TLYY266 pKa = 10.89AGKK269 pKa = 10.46SNAWIFEE276 pKa = 3.89FDD278 pKa = 3.02ITTPLDD284 pKa = 3.49PAFLGQFRR292 pKa = 11.84YY293 pKa = 8.49STGTTNQITDD303 pKa = 3.09NGTNMFIAFGIDD315 pKa = 3.2GLATAEE321 pKa = 4.14MQPMDD326 pKa = 4.37KK327 pKa = 10.26LDD329 pKa = 3.97HH330 pKa = 6.39FLVSILPNDD339 pKa = 3.68LSMDD343 pKa = 3.56NGRR346 pKa = 11.84LLASDD351 pKa = 3.58EE352 pKa = 4.33TGLFHH357 pKa = 7.9VIDD360 pKa = 4.24VEE362 pKa = 4.24NSFIPKK368 pKa = 9.88SRR370 pKa = 11.84IPYY373 pKa = 9.49ISNTFSGEE381 pKa = 3.89IKK383 pKa = 11.07NNTAWLGTGIYY394 pKa = 10.37LEE396 pKa = 4.85TIDD399 pKa = 6.22LSTLNPSSQIDD410 pKa = 3.65IQTPDD415 pKa = 2.93ASAEE419 pKa = 3.93VTYY422 pKa = 10.86LNEE425 pKa = 4.97IGDD428 pKa = 4.14TLYY431 pKa = 11.24LGTSSGMVAMYY442 pKa = 9.96DD443 pKa = 3.3VSGNVPSLIASITFPVHH460 pKa = 7.09AEE462 pKa = 4.22TNSPQYY468 pKa = 9.3ITDD471 pKa = 3.48VVPYY475 pKa = 10.4GDD477 pKa = 4.17YY478 pKa = 11.3LLASSIEE485 pKa = 4.15SEE487 pKa = 4.33LLIADD492 pKa = 4.77FSNLASPAVVNVPEE506 pKa = 4.22LSEE509 pKa = 3.96VAEE512 pKa = 4.4ANLFVTGDD520 pKa = 3.77KK521 pKa = 10.8LLSASDD527 pKa = 3.13WGAFLIDD534 pKa = 3.99ISDD537 pKa = 3.89IEE539 pKa = 4.22NPTYY543 pKa = 10.89YY544 pKa = 9.68GTQLVEE550 pKa = 4.34LGLITSATRR559 pKa = 11.84LDD561 pKa = 3.84ADD563 pKa = 3.64QVLLSSTEE571 pKa = 3.84GLLTVNIADD580 pKa = 4.65PDD582 pKa = 4.14NITIVSKK589 pKa = 10.86IEE591 pKa = 3.9SAPEE595 pKa = 3.81FSSVATDD602 pKa = 4.13GSHH605 pKa = 6.07VVGALHH611 pKa = 6.64YY612 pKa = 9.88EE613 pKa = 4.38SEE615 pKa = 4.21LKK617 pKa = 10.5LYY619 pKa = 10.36KK620 pKa = 10.2FNKK623 pKa = 10.14SPTTSDD629 pKa = 3.54YY630 pKa = 11.16EE631 pKa = 4.17FSVDD635 pKa = 3.37EE636 pKa = 4.72DD637 pKa = 4.18SSIVEE642 pKa = 4.26YY643 pKa = 11.4VEE645 pKa = 4.62ATDD648 pKa = 3.98PDD650 pKa = 3.62GDD652 pKa = 4.09EE653 pKa = 4.05IVFSIVSQPDD663 pKa = 3.27SGSVSITSAGQFTYY677 pKa = 10.79EE678 pKa = 4.36PNADD682 pKa = 3.81FNGQDD687 pKa = 2.88TFTFEE692 pKa = 5.73AEE694 pKa = 3.96DD695 pKa = 4.21TYY697 pKa = 11.62GGSSEE702 pKa = 4.22GSVSITINSVNDD714 pKa = 3.28APTASSVSLTTTVDD728 pKa = 3.27KK729 pKa = 11.08AVSGSFNASDD739 pKa = 3.53VDD741 pKa = 3.96GDD743 pKa = 3.69EE744 pKa = 4.16LTYY747 pKa = 11.28EE748 pKa = 4.11NTNPSNGSLSVSGSGFTYY766 pKa = 9.99TPAAGFSGQDD776 pKa = 2.99SFQYY780 pKa = 10.43TVTDD784 pKa = 3.25SGGEE788 pKa = 4.16SVTANVTITVNKK800 pKa = 8.43PASSGGGGGSSDD812 pKa = 3.67LWLLALLLLGFTFRR826 pKa = 11.84RR827 pKa = 11.84HH828 pKa = 4.52IRR830 pKa = 3.38
MM1 pKa = 7.37KK2 pKa = 10.36KK3 pKa = 10.32VIPFLASALAFLPQSVISQDD23 pKa = 3.6TILVDD28 pKa = 3.12TGEE31 pKa = 4.59VYY33 pKa = 10.94GHH35 pKa = 6.17GPYY38 pKa = 10.5YY39 pKa = 10.62GVEE42 pKa = 4.13AYY44 pKa = 10.56GNASYY49 pKa = 11.76VNDD52 pKa = 3.92GSKK55 pKa = 10.15EE56 pKa = 3.79IKK58 pKa = 10.76VFDD61 pKa = 3.91VSDD64 pKa = 3.69ASNITLSGEE73 pKa = 4.28VEE75 pKa = 4.43VGCTVDD81 pKa = 6.3DD82 pKa = 4.88LDD84 pKa = 6.25LDD86 pKa = 4.2GATLVAKK93 pKa = 10.36CPFEE97 pKa = 3.55IHH99 pKa = 6.81FYY101 pKa = 11.4DD102 pKa = 4.66LTDD105 pKa = 3.43SLNPEE110 pKa = 4.2LVGSYY115 pKa = 10.82DD116 pKa = 3.52STSYY120 pKa = 11.18AVNSVEE126 pKa = 4.59LSGDD130 pKa = 3.08RR131 pKa = 11.84LYY133 pKa = 10.63IGGGNTDD140 pKa = 2.58IVIFNASDD148 pKa = 3.57LSNIQEE154 pKa = 4.47LNSQTFDD161 pKa = 3.64SISTGEE167 pKa = 4.21LKK169 pKa = 10.65KK170 pKa = 10.76SANFVYY176 pKa = 10.45SISDD180 pKa = 3.54FDD182 pKa = 3.91TVKK185 pKa = 10.59IFDD188 pKa = 3.67VSNEE192 pKa = 3.68EE193 pKa = 4.35NIIKK197 pKa = 9.71TSEE200 pKa = 3.79INATATLFKK209 pKa = 10.8DD210 pKa = 3.42VLIIDD215 pKa = 3.64NTLYY219 pKa = 10.41IAATDD224 pKa = 3.67GLRR227 pKa = 11.84VYY229 pKa = 10.63DD230 pKa = 4.12VTNMASPSYY239 pKa = 11.0VKK241 pKa = 10.08TIDD244 pKa = 3.17SGSFFNTIDD253 pKa = 4.02LLNALHH259 pKa = 6.45VVGDD263 pKa = 3.49TLYY266 pKa = 10.89AGKK269 pKa = 10.46SNAWIFEE276 pKa = 3.89FDD278 pKa = 3.02ITTPLDD284 pKa = 3.49PAFLGQFRR292 pKa = 11.84YY293 pKa = 8.49STGTTNQITDD303 pKa = 3.09NGTNMFIAFGIDD315 pKa = 3.2GLATAEE321 pKa = 4.14MQPMDD326 pKa = 4.37KK327 pKa = 10.26LDD329 pKa = 3.97HH330 pKa = 6.39FLVSILPNDD339 pKa = 3.68LSMDD343 pKa = 3.56NGRR346 pKa = 11.84LLASDD351 pKa = 3.58EE352 pKa = 4.33TGLFHH357 pKa = 7.9VIDD360 pKa = 4.24VEE362 pKa = 4.24NSFIPKK368 pKa = 9.88SRR370 pKa = 11.84IPYY373 pKa = 9.49ISNTFSGEE381 pKa = 3.89IKK383 pKa = 11.07NNTAWLGTGIYY394 pKa = 10.37LEE396 pKa = 4.85TIDD399 pKa = 6.22LSTLNPSSQIDD410 pKa = 3.65IQTPDD415 pKa = 2.93ASAEE419 pKa = 3.93VTYY422 pKa = 10.86LNEE425 pKa = 4.97IGDD428 pKa = 4.14TLYY431 pKa = 11.24LGTSSGMVAMYY442 pKa = 9.96DD443 pKa = 3.3VSGNVPSLIASITFPVHH460 pKa = 7.09AEE462 pKa = 4.22TNSPQYY468 pKa = 9.3ITDD471 pKa = 3.48VVPYY475 pKa = 10.4GDD477 pKa = 4.17YY478 pKa = 11.3LLASSIEE485 pKa = 4.15SEE487 pKa = 4.33LLIADD492 pKa = 4.77FSNLASPAVVNVPEE506 pKa = 4.22LSEE509 pKa = 3.96VAEE512 pKa = 4.4ANLFVTGDD520 pKa = 3.77KK521 pKa = 10.8LLSASDD527 pKa = 3.13WGAFLIDD534 pKa = 3.99ISDD537 pKa = 3.89IEE539 pKa = 4.22NPTYY543 pKa = 10.89YY544 pKa = 9.68GTQLVEE550 pKa = 4.34LGLITSATRR559 pKa = 11.84LDD561 pKa = 3.84ADD563 pKa = 3.64QVLLSSTEE571 pKa = 3.84GLLTVNIADD580 pKa = 4.65PDD582 pKa = 4.14NITIVSKK589 pKa = 10.86IEE591 pKa = 3.9SAPEE595 pKa = 3.81FSSVATDD602 pKa = 4.13GSHH605 pKa = 6.07VVGALHH611 pKa = 6.64YY612 pKa = 9.88EE613 pKa = 4.38SEE615 pKa = 4.21LKK617 pKa = 10.5LYY619 pKa = 10.36KK620 pKa = 10.2FNKK623 pKa = 10.14SPTTSDD629 pKa = 3.54YY630 pKa = 11.16EE631 pKa = 4.17FSVDD635 pKa = 3.37EE636 pKa = 4.72DD637 pKa = 4.18SSIVEE642 pKa = 4.26YY643 pKa = 11.4VEE645 pKa = 4.62ATDD648 pKa = 3.98PDD650 pKa = 3.62GDD652 pKa = 4.09EE653 pKa = 4.05IVFSIVSQPDD663 pKa = 3.27SGSVSITSAGQFTYY677 pKa = 10.79EE678 pKa = 4.36PNADD682 pKa = 3.81FNGQDD687 pKa = 2.88TFTFEE692 pKa = 5.73AEE694 pKa = 3.96DD695 pKa = 4.21TYY697 pKa = 11.62GGSSEE702 pKa = 4.22GSVSITINSVNDD714 pKa = 3.28APTASSVSLTTTVDD728 pKa = 3.27KK729 pKa = 11.08AVSGSFNASDD739 pKa = 3.53VDD741 pKa = 3.96GDD743 pKa = 3.69EE744 pKa = 4.16LTYY747 pKa = 11.28EE748 pKa = 4.11NTNPSNGSLSVSGSGFTYY766 pKa = 9.99TPAAGFSGQDD776 pKa = 2.99SFQYY780 pKa = 10.43TVTDD784 pKa = 3.25SGGEE788 pKa = 4.16SVTANVTITVNKK800 pKa = 8.43PASSGGGGGSSDD812 pKa = 3.67LWLLALLLLGFTFRR826 pKa = 11.84RR827 pKa = 11.84HH828 pKa = 4.52IRR830 pKa = 3.38
Molecular weight: 88.74 kDa
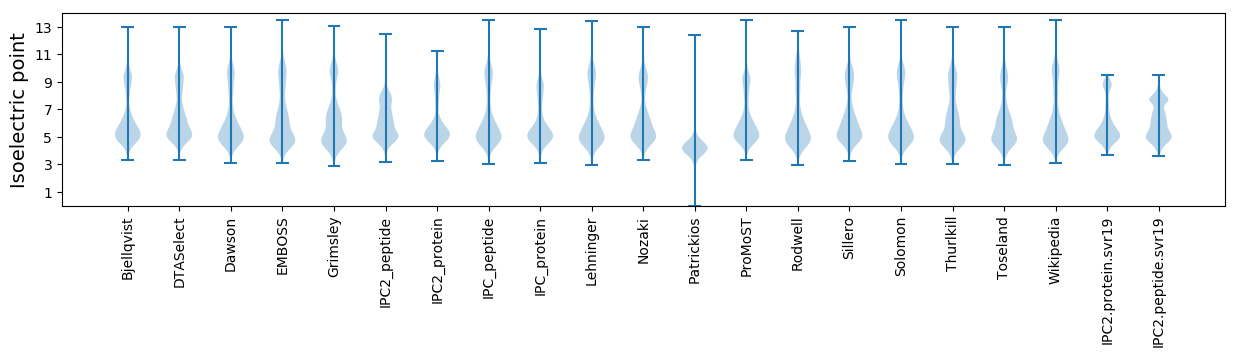
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7RAG2|C7RAG2_KANKD Uncharacterized protein OS=Kangiella koreensis (strain DSM 16069 / KCTC 12182 / SW-125) OX=523791 GN=Kkor_0834 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.53RR3 pKa = 11.84TFQPSIIKK11 pKa = 10.36RR12 pKa = 11.84KK13 pKa = 7.45RR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84QVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
MM1 pKa = 7.45KK2 pKa = 9.53RR3 pKa = 11.84TFQPSIIKK11 pKa = 10.36RR12 pKa = 11.84KK13 pKa = 7.45RR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84QVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
852378 |
30 |
2213 |
324.1 |
36.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.079 ± 0.048 | 0.89 ± 0.014 |
5.778 ± 0.037 | 6.798 ± 0.052 |
4.187 ± 0.033 | 6.815 ± 0.044 |
2.236 ± 0.025 | 6.621 ± 0.044 |
5.745 ± 0.04 | 10.195 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.498 ± 0.027 | 4.264 ± 0.036 |
3.941 ± 0.031 | 4.581 ± 0.042 |
4.442 ± 0.036 | 6.551 ± 0.041 |
5.082 ± 0.031 | 6.754 ± 0.034 |
1.301 ± 0.019 | 3.243 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
